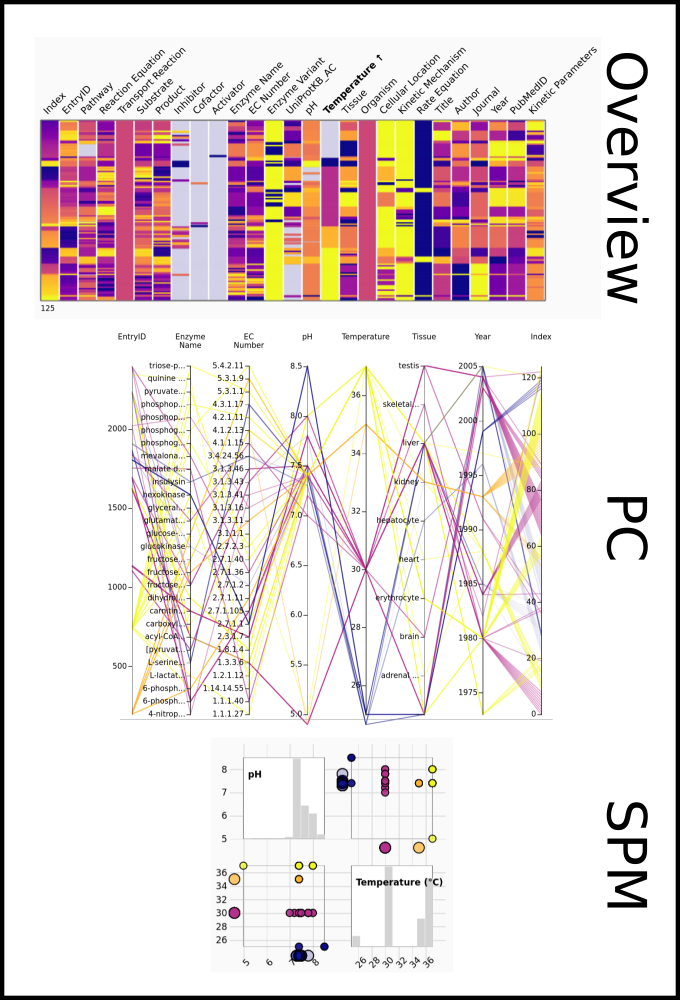
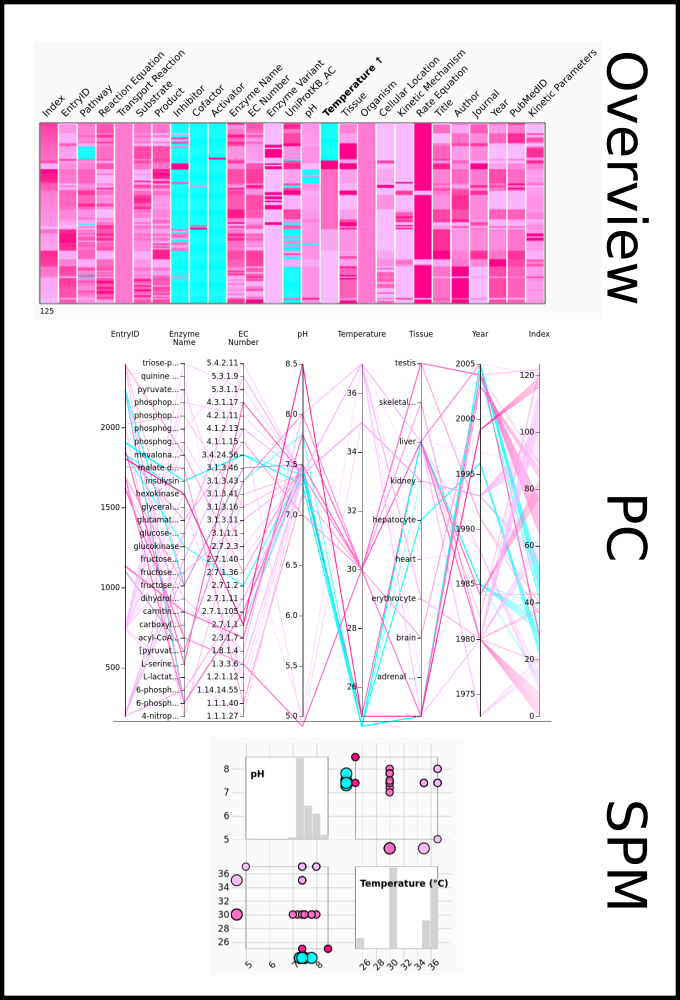
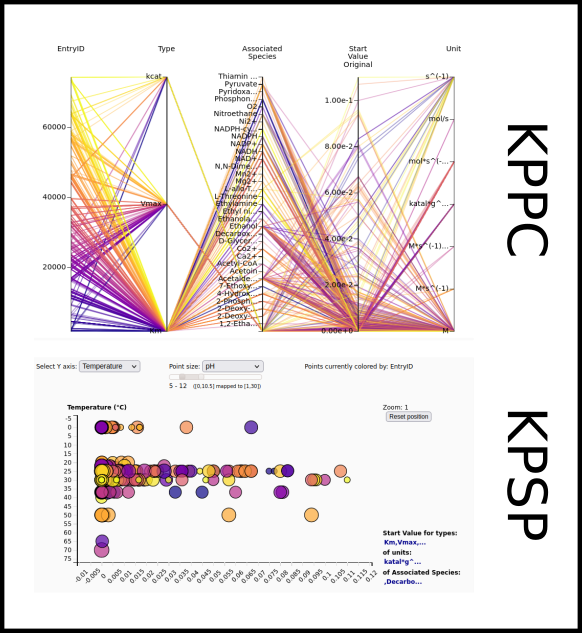
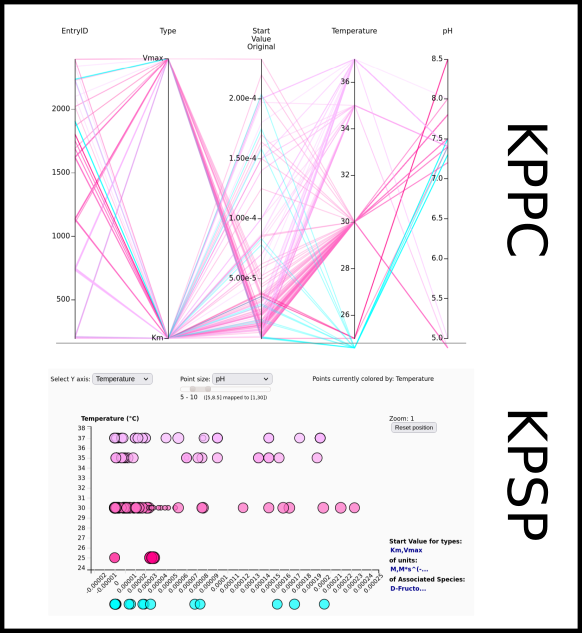
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
59001
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59002
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59003
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59004
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59005
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59006
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59007
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59008
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59009
|
MJ0109 is an enzyme that is both an inositol monophosphatase and the missing archaeal ...
|
|
59010
|
Pyrophosphate-dependent phosphofructokinase from the amoeba Naegleria fowleri, an AMP-sensitive enzyme.
|
|
59011
|
Pyrophosphate-dependent phosphofructokinase from the amoeba Naegleria fowleri, an AMP-sensitive enzyme.
|
|
59012
|
Pyrophosphate-dependent phosphofructokinase from the amoeba Naegleria fowleri, an AMP-sensitive enzyme.
|
|
59013
|
Pyrophosphate-dependent phosphofructokinase from the amoeba Naegleria fowleri, an AMP-sensitive enzyme.
|
|
59014
|
Pyrophosphate-dependent phosphofructokinase from the amoeba Naegleria fowleri, an AMP-sensitive enzyme.
|
|
59015
|
Pyrophosphate-dependent phosphofructokinase from the amoeba Naegleria fowleri, an AMP-sensitive enzyme.
|
|
59016
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59017
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59018
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59019
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59020
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59021
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59022
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59023
|
Ribonucleases with angiogenic and bactericidal activities from the Atlantic salmon.
|
|
59024
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59025
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59026
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59027
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59028
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59029
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59030
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59031
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59032
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59033
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59034
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59035
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59036
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59037
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59038
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59039
|
A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the ...
|
|
59040
|
Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. ...
|
|
59041
|
Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. ...
|
|
59042
|
Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. ...
|
|
59043
|
Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. ...
|
|
59044
|
Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. ...
|
|
59045
|
Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. ...
|
|
59046
|
Development of a mutagenesis, expression and purification system for yeast phosphoglycerate mutase. ...
|
|
59047
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59048
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59049
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59050
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59051
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59052
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59053
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59054
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59055
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59056
|
A specialized citric acid cycle requiring succinyl-coenzyme A (CoA):acetate CoA-transferase (AarC) confers ...
|
|
59057
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59058
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59059
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59060
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59061
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59062
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59063
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59064
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59065
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59066
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59067
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59068
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59069
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59070
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59071
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59072
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59073
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59074
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59075
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59076
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59077
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59078
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59079
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59080
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59081
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59082
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59083
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59084
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59085
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59086
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59087
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59088
|
Crystal structures of Acetobacter aceti succinyl-coenzyme A (CoA):acetate CoA-transferase reveal specificity ...
|
|
59089
|
Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison ...
|
|
59090
|
Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison ...
|
|
59091
|
Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison ...
|
|
59092
|
Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison ...
|
|
59093
|
Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison ...
|
|
59094
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59095
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59096
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59097
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59098
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59099
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59100
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59101
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59102
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59103
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59104
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59105
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59106
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59107
|
Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize.
|
|
59108
|
Fumarase a from Escherichia coli: purification and characterization as an iron-sulfur cluster containing ...
|
|
59109
|
Fumarase a from Escherichia coli: purification and characterization as an iron-sulfur cluster containing ...
|
|
59110
|
Fumarase a from Escherichia coli: purification and characterization as an iron-sulfur cluster containing ...
|
|
59111
|
Fumarase a from Escherichia coli: purification and characterization as an iron-sulfur cluster containing ...
|
|
59112
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59113
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59114
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59115
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59116
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59117
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59118
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59119
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59120
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59121
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59122
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59123
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59124
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59125
|
Purification and characterization of a beta-glucosidase from rye (Secale cereale L.) seedlings
|
|
59126
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59127
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59128
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59129
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59130
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59131
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59132
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59133
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59134
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59135
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59136
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59137
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59138
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59139
|
Purification and characterization of a hydroxamic acid glucoside beta-glucosidase from wheat (Triticum ...
|
|
59140
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59141
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59142
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59143
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59144
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59145
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59146
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59147
|
Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol ...
|
|
59148
|
Trans-membrane uptake and intracellular metabolism of fatty acids in Atlantic salmon (Salmo salar L.) ...
|
|
59149
|
Structure and properties of an engineered transketolase from maize.
|
|
59150
|
Puromycin-sensitive alanyl aminopeptidase from human liver cytosol: purification and characterization.
|
|
59151
|
Puromycin-sensitive alanyl aminopeptidase from human liver cytosol: purification and characterization.
|
|
59152
|
Puromycin-sensitive alanyl aminopeptidase from human liver cytosol: purification and characterization.
|
|
59153
|
Puromycin-sensitive alanyl aminopeptidase from human liver cytosol: purification and characterization.
|
|
59154
|
Puromycin-sensitive alanyl aminopeptidase from human liver cytosol: purification and characterization.
|
|
59155
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59156
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59157
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59158
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59159
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59160
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59161
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59162
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59163
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59164
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59165
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59166
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59167
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59168
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59169
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59170
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59171
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59172
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59173
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59174
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59175
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59176
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59177
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59178
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59179
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59180
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59181
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59182
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59183
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59184
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59185
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59186
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59187
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59188
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59189
|
Comparative study of substrate-inhibitor specificity of brain cholinesterase activity of pacific salmon fish ...
|
|
59190
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59191
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59192
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59193
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59194
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59195
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59196
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59197
|
Temperature and Ca(2+)-dependence of the sarcoplasmic reticulum Ca(2+)-ATPase in haddock, salmon, rainbow ...
|
|
59198
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59199
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59200
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59201
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59202
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59203
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59204
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59205
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59206
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59207
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59208
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59209
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59210
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59211
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59212
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59213
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59214
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59215
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59216
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59217
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59218
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59219
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59220
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59221
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59222
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59223
|
Purification and properties of the hepatic glutathione S-transferases of the Atlantic salmon (Salmo salar).
|
|
59224
|
Sucrose fermentation by Fusobacterium mortiferum ATCC 25557: transport, catabolism, and products
|
|
59225
|
Sucrose fermentation by Fusobacterium mortiferum ATCC 25557: transport, catabolism, and products
|
|
59226
|
Sucrose fermentation by Fusobacterium mortiferum ATCC 25557: transport, catabolism, and products
|
|
59227
|
Sucrose fermentation by Fusobacterium mortiferum ATCC 25557: transport, catabolism, and products
|
|
59228
|
Sucrose fermentation by Fusobacterium mortiferum ATCC 25557: transport, catabolism, and products
|
|
59229
|
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 Kinase that is activated when ...
|
|
59230
|
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 Kinase that is activated when ...
|
|
59231
|
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 Kinase that is activated when ...
|
|
59232
|
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 Kinase that is activated when ...
|
|
59233
|
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 Kinase that is activated when ...
|
|
59234
|
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 Kinase that is activated when ...
|
|
59235
|
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 Kinase that is activated when ...
|
|
59236
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59237
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59238
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59239
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59240
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59241
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59242
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59243
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59244
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59245
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59246
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59247
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59248
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59249
|
Structure of D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase bifunctional protein
|
|
59250
|
Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the ...
|
|
59251
|
Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the ...
|
|
59252
|
Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the ...
|
|
59253
|
Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the ...
|
|
59254
|
Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the ...
|
|
59255
|
Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the ...
|
|
59256
|
Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the ...
|
|
59257
|
Biochemical and genetic analysis of the four DNA ligases of mycobacteria.
|
|
59258
|
Biochemical and genetic analysis of the four DNA ligases of mycobacteria.
|
|
59259
|
Biochemical and genetic analysis of the four DNA ligases of mycobacteria.
|
|
59260
|
The Inhibition of Isocitrate Lyase from Escherichia coli by Glyoxylate
|
|
59261
|
The Inhibition of Isocitrate Lyase from Escherichia coli by Glyoxylate
|
|
59262
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59263
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59264
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59265
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59266
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59267
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59268
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59269
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59270
|
6-phosphofructokinase (pyrophosphate). Properties of the enzyme from Entamoeba histolytica and its reaction ...
|
|
59271
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59272
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59273
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59274
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59275
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59276
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59277
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59278
|
The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for ...
|
|
59279
|
A novel GDP-mannose mannosyl hydrolase shares homology with the MutT family of enzymes
|
|
59280
|
A novel GDP-mannose mannosyl hydrolase shares homology with the MutT family of enzymes
|
|
59281
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59282
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59283
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59284
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59285
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59286
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59287
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59288
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59289
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59290
|
Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast ...
|
|
59291
|
Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites.
|
|
59292
|
Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites.
|
|
59293
|
Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites.
|
|
59294
|
Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites.
|
|
59295
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59296
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59297
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59298
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59299
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59300
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59301
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59302
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59303
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59304
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59305
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59306
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59307
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59308
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59309
|
Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides ...
|
|
59310
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59311
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59312
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59313
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59314
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59315
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59316
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59317
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59318
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59319
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59320
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59321
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59322
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59323
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59324
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59325
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59326
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59327
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59328
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59329
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59330
|
Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and ...
|
|
59331
|
The active centers of adenylylsulfate reductase from Desulfovibrio gigas. Characterization and spectroscopic ...
|
|
59332
|
The active centers of adenylylsulfate reductase from Desulfovibrio gigas. Characterization and spectroscopic ...
|
|
59333
|
Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT
|
|
59334
|
Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT
|
|
59335
|
Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa)
|
|
59336
|
The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid ...
|
|
59337
|
The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid ...
|
|
59338
|
The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid ...
|
|
59339
|
The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid ...
|
|
59340
|
The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid ...
|
|
59341
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59342
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59343
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59344
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59345
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59346
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59347
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59348
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59349
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59350
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59351
|
Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme
|
|
59352
|
Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis
|
|
59353
|
Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis
|
|
59354
|
Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis
|
|
59355
|
Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis
|
|
59356
|
Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis
|
|
59357
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59358
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59359
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59360
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59361
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59362
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59363
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59364
|
Molecular identification and characterization of novel human and mouse concentrative Na+-nucleoside ...
|
|
59365
|
Identification of thermostable glyoxalase I in the fission yeast Schizosaccharomyces pombe.
|
|
59366
|
Identification of thermostable glyoxalase I in the fission yeast Schizosaccharomyces pombe.
|
|
59367
|
Catalytic mechanism of retaining alpha-galactosidase belonging to glycoside hydrolase family 97.
|
|
59368
|
Catalytic mechanism of retaining alpha-galactosidase belonging to glycoside hydrolase family 97.
|
|
59369
|
Catalytic mechanism of retaining alpha-galactosidase belonging to glycoside hydrolase family 97.
|
|
59370
|
Catalytic mechanism of retaining alpha-galactosidase belonging to glycoside hydrolase family 97.
|
|
59371
|
The presence of two GDP-L-fucose: glycoproteine fucosyltransferases in human serum
|
|
59372
|
The presence of two GDP-L-fucose: glycoproteine fucosyltransferases in human serum
|
|
59373
|
The presence of two GDP-L-fucose: glycoproteine fucosyltransferases in human serum
|
|
59374
|
The presence of two GDP-L-fucose: glycoproteine fucosyltransferases in human serum
|
|
59375
|
A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis
|
|
59376
|
A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis
|
|
59377
|
Characterization of recombinant Dictyostelium discoideum sepiapterin reductase expressed in E. coli.
|
|
59378
|
Orf135 from Escherichia coli Is a Nudix hydrolase specific for CTP, dCTP, and 5-methyl-dCTP.
|
|
59379
|
Orf135 from Escherichia coli Is a Nudix hydrolase specific for CTP, dCTP, and 5-methyl-dCTP.
|
|
59380
|
Orf135 from Escherichia coli Is a Nudix hydrolase specific for CTP, dCTP, and 5-methyl-dCTP.
|
|
59381
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59382
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59383
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59384
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59385
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59386
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59387
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59388
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59389
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59390
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59391
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59392
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59393
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59394
|
Natural substrates and inhibitors of mannan-binding lectin-associated serine protease-1 and -2: a study on ...
|
|
59395
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59396
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59397
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59398
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59399
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59400
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59401
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59402
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59403
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59404
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59405
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59406
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59407
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59408
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59409
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59410
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59411
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59412
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59413
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59414
|
Kinetic mechanisms of polyphosphate glucokinase from Mycobacterium tuberculosis.
|
|
59415
|
A novel 4-hydroxycoumarin biosynthetic pathway.
|
|
59416
|
A novel 4-hydroxycoumarin biosynthetic pathway.
|
|
59417
|
A novel 4-hydroxycoumarin biosynthetic pathway.
|
|
59418
|
A novel 4-hydroxycoumarin biosynthetic pathway.
|
|
59419
|
A novel 4-hydroxycoumarin biosynthetic pathway.
|
|
59420
|
A novel 4-hydroxycoumarin biosynthetic pathway.
|
|
59421
|
FlaA1, a new bifunctional UDP-GlcNAc C6 Dehydratase/ C4 reductase from Helicobacter pylori.
|
|
59422
|
FlaA1, a new bifunctional UDP-GlcNAc C6 Dehydratase/ C4 reductase from Helicobacter pylori.
|
|
59423
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59424
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59425
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59426
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59427
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59428
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59429
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59430
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59431
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59432
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59433
|
Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A
|
|
59434
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59435
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59436
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59437
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59438
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59439
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59440
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59441
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59442
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59443
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59444
|
Stereochemical course and steady state mechanism of the reaction catalyzed by the GDP-fucose synthetase from ...
|
|
59445
|
Inducible membrane-bound L-lactate dehydrogenase from Escherichia coli. Purification and properties.
|
|
59446
|
Inducible membrane-bound L-lactate dehydrogenase from Escherichia coli. Purification and properties.
|
|
59447
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59448
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59449
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59450
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59451
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59452
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59453
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59454
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59455
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59456
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59457
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59458
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59459
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59460
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59461
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59462
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59463
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59464
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59465
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59466
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59467
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59468
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59469
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59470
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59471
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59472
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59473
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59474
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59475
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59476
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59477
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59478
|
Triacylglycerol-, wax ester- and sterol ester-hydrolases in midgut of Atlantic salmon (Salmo salar)
|
|
59479
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59480
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59481
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59482
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59483
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59484
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59485
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59486
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59487
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59488
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59489
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59490
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59491
|
The Cdc25 protein of Saccharomyces cerevisiae is required for normal glucose transport
|
|
59492
|
Cloning and expression of a glycine transporter reveal colocalization with NMDA receptors.
|
|
59493
|
Cloning and expression of a glycine transporter reveal colocalization with NMDA receptors.
|
|
59494
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59495
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59496
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59497
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59498
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59499
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59500
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59501
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59502
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59503
|
The nature of the catalytic domain of 2'-5'-oligoadenylate synthetases.
|
|
59504
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59505
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59506
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59507
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59508
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59509
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59510
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59511
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59512
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59513
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59514
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59515
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59516
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59517
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59518
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59519
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59520
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59521
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59522
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59523
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59524
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59525
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59526
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59527
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59528
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59529
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59530
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59531
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59532
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59533
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59534
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59535
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59536
|
Molecular characterization of carbamoyl-phosphate synthetase (CPS1) deficiency using human recombinant CPS1 as ...
|
|
59537
|
Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1
|
|
59538
|
Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1
|
|
59539
|
Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1
|
|
59540
|
Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1
|
|
59541
|
Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1
|
|
59542
|
Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1
|
|
59543
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59544
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59545
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59546
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59547
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59548
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59549
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59550
|
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, ...
|
|
59551
|
Stilbene synthase from Scots pine (Pinus sylvestris).
|
|
59552
|
Stilbene synthase from Scots pine (Pinus sylvestris).
|
|
59553
|
Purification and characterization of malate synthase from the glucose-grown wood-rotting basidiomycete ...
|
|
59554
|
Purification and characterization of malate synthase from the glucose-grown wood-rotting basidiomycete ...
|
|
59555
|
Purification and characterization of malate synthase from the glucose-grown wood-rotting basidiomycete ...
|
|
59556
|
Purification and characterization of malate synthase from the glucose-grown wood-rotting basidiomycete ...
|
|
59557
|
Purification and characterization of malate synthase from the glucose-grown wood-rotting basidiomycete ...
|
|
59558
|
Purification and characterization of malate synthase from the glucose-grown wood-rotting basidiomycete ...
|
|
59559
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59560
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59561
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59562
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59563
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59564
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59565
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59566
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59567
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59568
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59569
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59570
|
Role of highly conserved residues in the reaction catalyzed by recombinant Delta7-sterol-C5(6)-desaturase ...
|
|
59571
|
Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic ...
|
|
59572
|
Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic ...
|
|
59573
|
Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic ...
|
|
59574
|
Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic ...
|
|
59575
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59576
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59577
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59578
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59579
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59580
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59581
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59582
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59583
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59584
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59585
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59586
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59587
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59588
|
Striatum- and testis-specific phosphodiesterase PDE10A isolation and characterization of a rat PDE10A
|
|
59589
|
Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis
|
|
59590
|
Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis
|
|
59591
|
Purification and characterization of a platelet aggregation inhibitor acidic phospholipase A2 from Indian ...
|
|
59592
|
Expression, characterization, and crystallization of the pyrophosphate-dependent phosphofructo-1-kinase of ...
|
|
59593
|
Expression, characterization, and crystallization of the pyrophosphate-dependent phosphofructo-1-kinase of ...
|
|
59594
|
A non-specific aminopeptidase from Aspergillus
|
|
59595
|
A non-specific aminopeptidase from Aspergillus
|
|
59596
|
A non-specific aminopeptidase from Aspergillus
|
|
59597
|
A non-specific aminopeptidase from Aspergillus
|
|
59598
|
A non-specific aminopeptidase from Aspergillus
|
|
59599
|
A non-specific aminopeptidase from Aspergillus
|
|
59600
|
A non-specific aminopeptidase from Aspergillus
|
|
59601
|
Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis ...
|
|
59602
|
Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis ...
|
|
59603
|
Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis ...
|
|
59604
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59605
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59606
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59607
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59608
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59609
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59610
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59611
|
Purification, molecular cloning, and catalytic activity of Schizosaccharomyces pombe pyridoxal reductase. A ...
|
|
59612
|
Human fatty acid omega-hydroxylase, CYP4A11: determination of complete genomic sequence and characterization ...
|
|
59613
|
Biosynthesis of terpenoids: 4-diphosphocytidyl-2C-methyl-D-erythritol synthase of Arabidopsis thaliana
|
|
59614
|
Biosynthesis of terpenoids: 4-diphosphocytidyl-2C-methyl-D-erythritol synthase of Arabidopsis thaliana
|
|
59615
|
Molecular cloning and characterization of a gene encoding glutaminase from Aspergillus oryzae
|
|
59616
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59617
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59618
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59619
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59620
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59621
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59622
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59623
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59624
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59625
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59626
|
Purification and biochemical properties of an N-hydroxyarylamine O-acetyltransferase from Escherichia coli
|
|
59627
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59628
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59629
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59630
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59631
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59632
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59633
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59634
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59635
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59636
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59637
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59638
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59639
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59640
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59641
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59642
|
Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in ...
|
|
59643
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59644
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59645
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59646
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59647
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59648
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59649
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59650
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59651
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59652
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59653
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59654
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59655
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59656
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59657
|
Characterization of chondroitin sulfate lyase ABC from Bacteroides thetaiotaomicron WAL2926
|
|
59658
|
Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and ...
|
|
59659
|
Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and ...
|
|
59660
|
Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and ...
|
|
59661
|
Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and ...
|
|
59662
|
Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and ...
|
|
59663
|
Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and ...
|
|
59664
|
Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and ...
|
|
59665
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59666
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59667
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59668
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59669
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59670
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59671
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59672
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59673
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59674
|
One scaffold, three binding modes: novel and selective pteridine reductase 1 inhibitors derived from fragment ...
|
|
59675
|
Surface hydrolysis of sphingomyelin by the outer membrane protein Rv0888 supports replication of Mycobacterium ...
|
|
59676
|
YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids
|
|
59677
|
YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids
|
|
59678
|
YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids
|
|
59679
|
YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids
|
|
59680
|
A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase
|
|
59681
|
Inhibition of phosphate and arsenate uptake in yeast by monoiodoacetate, fluoride, 2,4-dinitrophenol and ...
|
|
59682
|
Inhibition of phosphate and arsenate uptake in yeast by monoiodoacetate, fluoride, 2,4-dinitrophenol and ...
|
|
59683
|
Inhibition of phosphate and arsenate uptake in yeast by monoiodoacetate, fluoride, 2,4-dinitrophenol and ...
|
|
59684
|
Inhibition of phosphate and arsenate uptake in yeast by monoiodoacetate, fluoride, 2,4-dinitrophenol and ...
|
|
59685
|
Factors governing substrate-induced generation and extrusion of protons in the yeast Saccharomyces cerevisiae.
|
|
59686
|
Factors governing substrate-induced generation and extrusion of protons in the yeast Saccharomyces cerevisiae.
|
|
59687
|
Factors governing substrate-induced generation and extrusion of protons in the yeast Saccharomyces cerevisiae.
|
|
59688
|
Factors governing substrate-induced generation and extrusion of protons in the yeast Saccharomyces cerevisiae.
|
|
59689
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59690
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59691
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59692
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59693
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59694
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59695
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59696
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59697
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59698
|
Conservation and Role of Electrostatics in Thymidylate Synthase
|
|
59699
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59700
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59701
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59702
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59703
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59704
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59705
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59706
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59707
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59708
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59709
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59710
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59711
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59712
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59713
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59714
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59715
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59716
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59717
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59718
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59719
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59720
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59721
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59722
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59723
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59724
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59725
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59726
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59727
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59728
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59729
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59730
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59731
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59732
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59733
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59734
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59735
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59736
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59737
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59738
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59739
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59740
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59741
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59742
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59743
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59744
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59745
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59746
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59747
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59748
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59749
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59750
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59751
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59752
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59753
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59754
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59755
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59756
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59757
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59758
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59759
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59760
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59761
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59762
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59763
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59764
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59765
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59766
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59767
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59768
|
Mapping the active site of meprin-A with peptide substrates and inhibitors.
|
|
59769
|
Cloning and characterization of a cysteine proteinase from Saccharomyces cerevisiae.
|
|
59770
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59771
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59772
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59773
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59774
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59775
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59776
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59777
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59778
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59779
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59780
|
Glutamate dehydrogenase from Escherichia coli: purification and properties.
|
|
59781
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59782
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59783
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59784
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59785
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59786
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59787
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59788
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59789
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59790
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59791
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59792
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59793
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59794
|
Tissue distribution and kinetic characteristics of rat steroid 5 alpha-reductase isozymes. Evidence for ...
|
|
59795
|
Glucose-induced activation of plasma membrane H(+)-ATPase in mutants of the yeast Saccharomyces cerevisiae ...
|
|
59796
|
Metabolism of 2-oxoaldehyde in yeasts. Purification and characterization of NADPH-dependent ...
|
|
59797
|
Metabolism of 2-oxoaldehyde in yeasts. Purification and characterization of NADPH-dependent ...
|
|
59798
|
Metabolism of 2-oxoaldehyde in yeasts. Purification and characterization of NADPH-dependent ...
|
|
59799
|
Metabolism of 2-oxoaldehyde in yeasts. Purification and characterization of NADPH-dependent ...
|
|
59800
|
Metabolism of 2-oxoaldehyde in yeasts. Purification and characterization of NADPH-dependent ...
|
|
59801
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59802
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59803
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59804
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59805
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59806
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59807
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59808
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59809
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59810
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59811
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59812
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59813
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59814
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59815
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59816
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59817
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59818
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59819
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59820
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59821
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59822
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59823
|
Biochemical characterization of the heteromeric Bacillus subtilis dihydroorotate dehydrogenase and its ...
|
|
59824
|
Cloning and expression of the chicken type 2 iodothyronine 5'-deiodinase
|
|
59825
|
Cloning and expression of the chicken type 2 iodothyronine 5'-deiodinase
|
|
59826
|
Cloning and expression of the chicken type 2 iodothyronine 5'-deiodinase
|
|
59827
|
Cloning and expression of the chicken type 2 iodothyronine 5'-deiodinase
|
|
59828
|
Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein
|
|
59829
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59830
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59831
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59832
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59833
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59834
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59835
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59836
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59837
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59838
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59839
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59840
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59841
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59842
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59843
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59844
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59845
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59846
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59847
|
Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase
|
|
59848
|
Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties.
|
|
59849
|
Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties.
|
|
59850
|
Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties.
|
|
59851
|
Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties.
|
|
59852
|
Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties.
|
|
59853
|
Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties.
|
|
59854
|
Creatinine amidohydrolase (creatininase) from Pseudomonas putida. Purification and some properties.
|
|
59855
|
Identification of a UDP-GlcNAc:Skp1-hydroxyproline GlcNAc-transferase in the cytoplasm of Dictyostelium
|
|
59856
|
Identification of a UDP-GlcNAc:Skp1-hydroxyproline GlcNAc-transferase in the cytoplasm of Dictyostelium
|
|
59857
|
Identification of a UDP-GlcNAc:Skp1-hydroxyproline GlcNAc-transferase in the cytoplasm of Dictyostelium
|
|
59858
|
Purification, characterization, and cDNA cloning of a novel acidic endoglycoceramidase from the jellyfish, ...
|
|
59859
|
Hydrolytic editing by a class II aminoacyl-tRNA synthetase
|
|
59860
|
Hydrolytic editing by a class II aminoacyl-tRNA synthetase
|
|
59861
|
Hydrolytic editing by a class II aminoacyl-tRNA synthetase
|
|
59862
|
Research note: kinetic and inhibition studies with turkey acrosin.
|
|
59863
|
Research note: kinetic and inhibition studies with turkey acrosin.
|
|
59864
|
Research note: kinetic and inhibition studies with turkey acrosin.
|
|
59865
|
Research note: kinetic and inhibition studies with turkey acrosin.
|
|
59866
|
Research note: kinetic and inhibition studies with turkey acrosin.
|
|
59867
|
Research note: kinetic and inhibition studies with turkey acrosin.
|
|
59868
|
Research note: kinetic and inhibition studies with turkey acrosin.
|
|
59869
|
Emodin O-methyltransferase from Aspergillus terreus.
|
|
59870
|
Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic ...
|
|
59871
|
Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic ...
|
|
59872
|
Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic ...
|
|
59873
|
Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic ...
|
|
59874
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59875
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59876
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59877
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59878
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59879
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59880
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59881
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59882
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59883
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59884
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59885
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59886
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59887
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59888
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59889
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59890
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59891
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59892
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59893
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59894
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59895
|
Mutant PTR1 proteins from Leishmania tarentolae: comparative kinetic properties and active-site labeling
|
|
59896
|
Somatostatin binding to hepatocytes isolated from rainbow trout, Oncorhynchus mykiss, is modulated by insulin ...
|
|
59897
|
Somatostatin binding to hepatocytes isolated from rainbow trout, Oncorhynchus mykiss, is modulated by insulin ...
|
|
59898
|
Cloning and characterization of long-chain fatty alcohol oxidase LjFAO1 in lotus japonicus
|
|
59899
|
Cloning and characterization of long-chain fatty alcohol oxidase LjFAO1 in lotus japonicus
|
|
59900
|
Cloning and characterization of long-chain fatty alcohol oxidase LjFAO1 in lotus japonicus
|
|
59901
|
Cloning and characterization of long-chain fatty alcohol oxidase LjFAO1 in lotus japonicus
|
|
59902
|
Cloning and characterization of long-chain fatty alcohol oxidase LjFAO1 in lotus japonicus
|
|
59903
|
Cloning and characterization of long-chain fatty alcohol oxidase LjFAO1 in lotus japonicus
|
|
59904
|
Identification of FAH domain-containing protein 1 (FAHD1) as oxaloacetate decarboxylase
|
|
59905
|
Identification of FAH domain-containing protein 1 (FAHD1) as oxaloacetate decarboxylase
|
|
59906
|
Identification of Human Fumarylacetoacetate Hydrolase Domain-containing Protein 1 (FAHD1) as a Novel ...
|
|
59907
|
Kinetic analysis of PPi-dependent phosphofructokinase from Porphyromonas gingivalis
|
|
59908
|
Kinetic analysis of PPi-dependent phosphofructokinase from Porphyromonas gingivalis
|
|
59909
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59910
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59911
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59912
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59913
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59914
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59915
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59916
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59917
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59918
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59919
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59920
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59921
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59922
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59923
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59924
|
Cloning and characterization of a novel human phosphodiesterase that hydrolyzes both cAMP and cGMP (PDE10A)
|
|
59925
|
Identification of the gene encoding sulfopyruvate decarboxylase, an enzyme involved in biosynthesis of ...
|
|
59926
|
Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with ...
|
|
59927
|
Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of ...
|
|
59928
|
Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of ...
|
|
59929
|
Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli
|
|
59930
|
Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli
|
|
59931
|
Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the ...
|
|
59932
|
Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the ...
|
|
59933
|
Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the ...
|
|
59934
|
Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the ...
|
|
59935
|
Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the ...
|
|
59936
|
Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the ...
|
|
59937
|
Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the ...
|
|
59938
|
Functional expression and characterisation of a new human phosphatidylinositol 4-kinase PI4K230
|
|
59939
|
Functional expression and characterisation of a new human phosphatidylinositol 4-kinase PI4K230
|
|
59940
|
Entry of ketoconazole into Candida albicans.
|
|
59941
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59942
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59943
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59944
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59945
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59946
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59947
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59948
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59949
|
Mutated forms of phosphoglycerate mutase in yeast affect reversal of metabolic flux. Effect of reversible and ...
|
|
59950
|
Biochemical and molecular biological evidence for the presence of type II iodothyronine deiodinase in mouse ...
|
|
59951
|
Biochemical and molecular biological evidence for the presence of type II iodothyronine deiodinase in mouse ...
|
|
59952
|
Isolation and characterization of a dinucleoside triphosphatase from Saccharomyces cerevisiae
|
|
59953
|
Isolation and characterization of a dinucleoside triphosphatase from Saccharomyces cerevisiae
|
|
59954
|
Studies on the microsomal electron-transport system of anaerobically grown yeast. IV. Purification and ...
|
|
59955
|
Studies on the microsomal electron-transport system of anaerobically grown yeast. IV. Purification and ...
|
|
59956
|
Studies on the microsomal electron-transport system of anaerobically grown yeast. IV. Purification and ...
|
|
59957
|
Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-D-manno-octulosonic acid ...
|
|
59958
|
Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-D-manno-octulosonic acid ...
|
|
59959
|
AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A
|
|
59960
|
AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A
|
|
59961
|
AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A
|
|
59962
|
AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A
|
|
59963
|
AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A
|
|
59964
|
AknK is an L-2-deoxyfucosyltransferase in the biosynthesis of the anthracycline aclacinomycin A
|
|
59965
|
Arsenate uptake and release in relation to the inhibition of transport and glycolysis in yeast.
|
|
59966
|
Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence ...
|
|
59967
|
Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence ...
|
|
59968
|
cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and ...
|
|
59969
|
cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and ...
|
|
59970
|
cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and ...
|
|
59971
|
cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and ...
|
|
59972
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59973
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59974
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59975
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59976
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59977
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59978
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59979
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59980
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59981
|
Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation
|
|
59982
|
The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic ...
|
|
59983
|
The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic ...
|
|
59984
|
The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic ...
|
|
59985
|
The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic ...
|
|
59986
|
The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic ...
|
|
59987
|
The antiviral nucleotide analogs cidofovir and adefovir are novel substrates for human and rat renal organic ...
|
|
59988
|
Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive ...
|
|
59989
|
The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its ...
|
|
59990
|
The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its ...
|
|
59991
|
The Saccharomyces cerevisiae PCD1 gene encodes a peroxisomal nudix hydrolase active toward coenzyme A and its ...
|
|
59992
|
Bakers' yeast uridine nucleosidase. Purification, composition, and physical and enzymatic properties.
|
|
59993
|
Bakers' yeast uridine nucleosidase. Purification, composition, and physical and enzymatic properties.
|
|
59994
|
Bakers' yeast uridine nucleosidase. Purification, composition, and physical and enzymatic properties.
|
|
59995
|
Bakers' yeast uridine nucleosidase. Purification, composition, and physical and enzymatic properties.
|
|
59996
|
Bakers' yeast uridine nucleosidase. Purification, composition, and physical and enzymatic properties.
|
|
59997
|
Two ribose-5-phosphate isomerases from Escherichia coli K12: partial characterisation of the enzymes and ...
|
|
59998
|
Two ribose-5-phosphate isomerases from Escherichia coli K12: partial characterisation of the enzymes and ...
|
|
59999
|
Two ribose-5-phosphate isomerases from Escherichia coli K12: partial characterisation of the enzymes and ...
|
|
60000
|
Two ribose-5-phosphate isomerases from Escherichia coli K12: partial characterisation of the enzymes and ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.