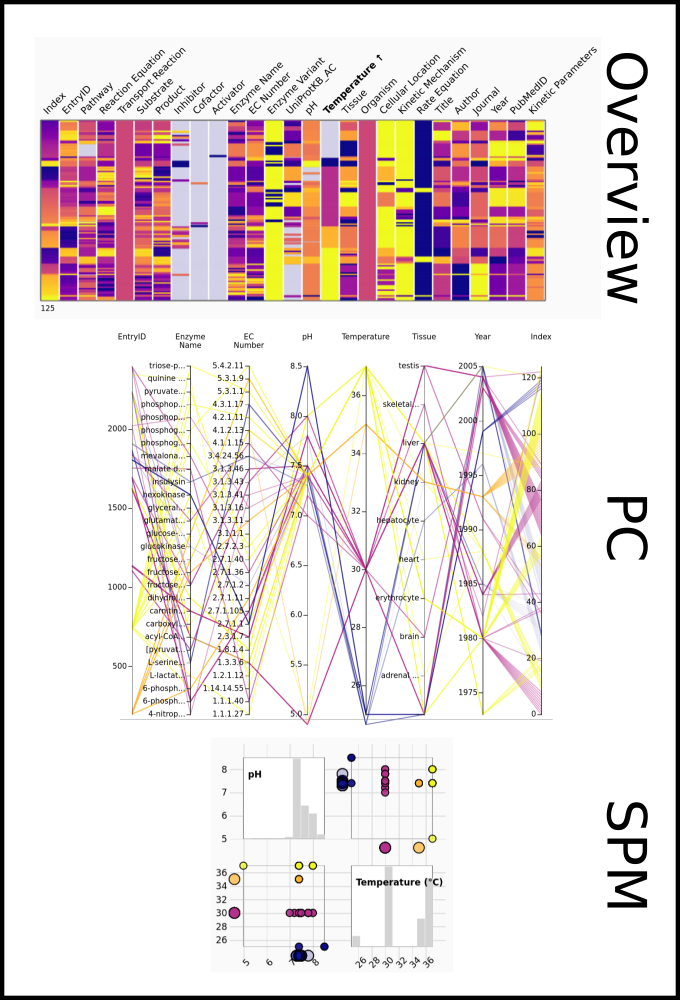
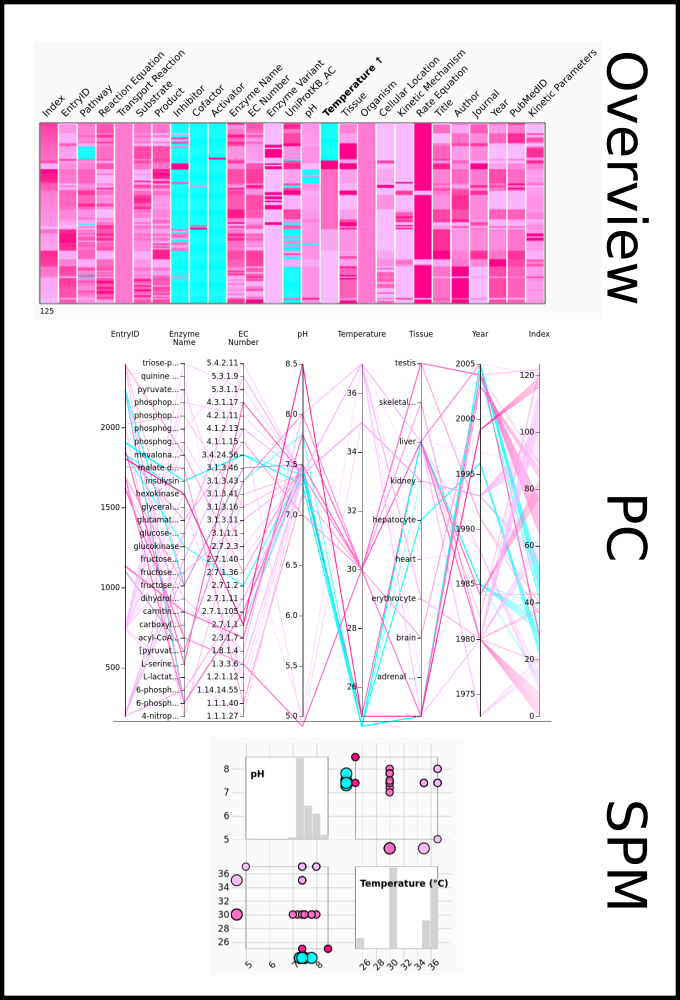
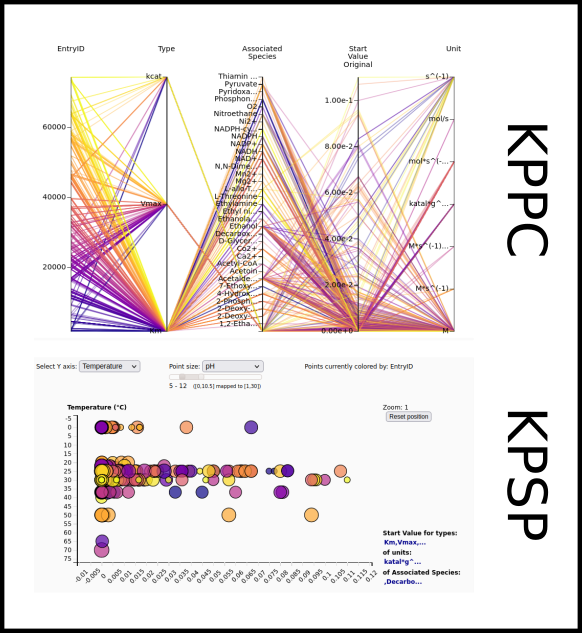
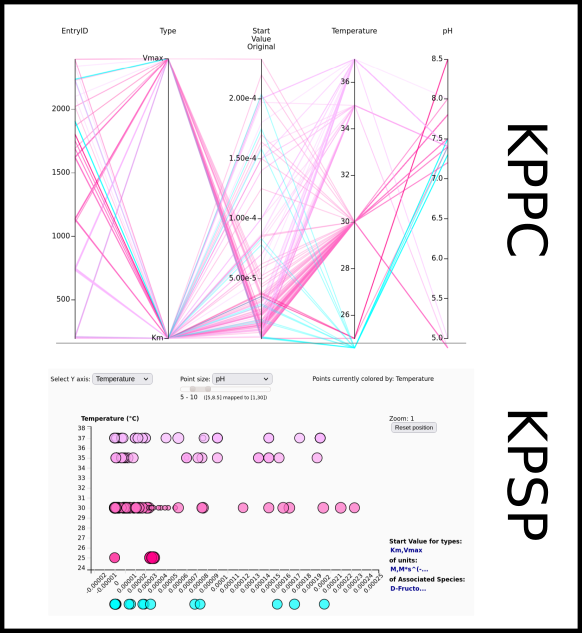
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
5001
|
Fluorinated aminoglycosides and their mechanistic implication for aminoglycoside 3`-phosphotransferases from ...
|
|
5002
|
Fluorinated aminoglycosides and their mechanistic implication for aminoglycoside 3`-phosphotransferases from ...
|
|
5003
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5004
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5005
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5006
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5007
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5008
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5009
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5010
|
Active site mutants of Drosophila melanogaster multisubstrate deoxyribonucleoside kinase
|
|
5011
|
The metabolism of nitrosothiols in the Mycobacteria: identification and characterization of S-nitrosomycothiol ...
|
|
5012
|
Mechanism of the reaction catalyzed by dehydroascorbate reductase from spinach chloroplasts
|
|
5013
|
Mechanism of the reaction catalyzed by dehydroascorbate reductase from spinach chloroplasts
|
|
5014
|
Mechanism of the reaction catalyzed by dehydroascorbate reductase from spinach chloroplasts
|
|
5015
|
Mechanism of the reaction catalyzed by dehydroascorbate reductase from spinach chloroplasts
|
|
5016
|
Catalysis and function of the p38 alpha.MK2a signaling complex
|
|
5017
|
Catalysis and function of the p38 alpha.MK2a signaling complex
|
|
5018
|
Catalysis and function of the p38 alpha.MK2a signaling complex
|
|
5019
|
Fatty Acid Synthetase
|
|
5020
|
Fatty Acid Synthetase
|
|
5021
|
Fatty Acid Synthetase
|
|
5022
|
Fatty Acid Synthetase
|
|
5023
|
Fatty Acid Synthetase
|
|
5024
|
Fatty Acid Synthetase
|
|
5025
|
Fatty Acid Synthetase
|
|
5026
|
Fatty Acid Synthetase
|
|
5027
|
Kinetic Studies on Pig Heart Cytoplasmic Malate Dehydrogenase
|
|
5028
|
Kinetic Studies on Pig Heart Cytoplasmic Malate Dehydrogenase
|
|
5029
|
Kinetic Studies on Pig Heart Cytoplasmic Malate Dehydrogenase
|
|
5030
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5031
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5032
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5033
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5034
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5035
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5036
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5037
|
Origin of Cooperativity in the Activation of Fructose-1,6-bisphosphatase by Mg2
|
|
5038
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5039
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5040
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5041
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5042
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5043
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5044
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5045
|
Structural and kinetic analysis of catalysis by a thiamin diphosphate-dependent enzyme, benzoylformate ...
|
|
5046
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5047
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5048
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5049
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5050
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5051
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5052
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5053
|
Kinetic properties of human thymidylate synthase, an anticancer drug target
|
|
5054
|
High-throughput screening for potent and selective inhibitors of Plasmodium falciparum dihydroorotate ...
|
|
5055
|
High-throughput screening for potent and selective inhibitors of Plasmodium falciparum dihydroorotate ...
|
|
5056
|
High-throughput screening for potent and selective inhibitors of Plasmodium falciparum dihydroorotate ...
|
|
5057
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5058
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5059
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5060
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5061
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5062
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5063
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5064
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5065
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5066
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5067
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5068
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5069
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5070
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5071
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5072
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5073
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5074
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5075
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5076
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5077
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5078
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5079
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5080
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5081
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5082
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5083
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5084
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5085
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5086
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5087
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5088
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5089
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5090
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5091
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5092
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5093
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5094
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5095
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5096
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5097
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5098
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5099
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5100
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5101
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5102
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5103
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5104
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5105
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5106
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5107
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5108
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5109
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5110
|
Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase
|
|
5111
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5112
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5113
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5114
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5115
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5116
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5117
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5118
|
Kinetic Analysis of the L-Ornithine transcarbamoylase from Pseudomonas savastanoi pv. phaseolicola That Is ...
|
|
5119
|
Biosynthesis reaction mechanism and kinetics of deoxynucleoside triphosphates, dATP and dGTP
|
|
5120
|
Biosynthesis reaction mechanism and kinetics of deoxynucleoside triphosphates, dATP and dGTP
|
|
5121
|
Kinetic evaluation of catalase and peroxygenase activities of tyrosinase
|
|
5122
|
Kinetic evaluation of catalase and peroxygenase activities of tyrosinase
|
|
5123
|
Kinetic evaluation of catalase and peroxygenase activities of tyrosinase
|
|
5124
|
Kinetic evaluation of catalase and peroxygenase activities of tyrosinase
|
|
5125
|
Kinetic evaluation of catalase and peroxygenase activities of tyrosinase
|
|
5126
|
Kinetic evaluation of catalase and peroxygenase activities of tyrosinase
|
|
5127
|
Kinetic evaluation of catalase and peroxygenase activities of tyrosinase
|
|
5128
|
RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein
|
|
5129
|
RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein
|
|
5130
|
RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein
|
|
5131
|
RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein
|
|
5132
|
RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein
|
|
5133
|
RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein
|
|
5134
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5135
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5136
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5137
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5138
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5139
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5140
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5141
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5142
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5143
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5144
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5145
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5146
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5147
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5148
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5149
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5150
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5151
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5152
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5153
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5154
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5155
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5156
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5157
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5158
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5159
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5160
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5161
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5162
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5163
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5164
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5165
|
Mechanistic features of lignin peroxidase-catalyzed oxidation of substituted phenols and 1,2-dimethoxyarenes
|
|
5166
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5167
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5168
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5169
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5170
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5171
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5172
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5173
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5174
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5175
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5176
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5177
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5178
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5179
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5180
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5181
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5182
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5183
|
Characterization of the ligandin site of maize glutathione S-transferase I
|
|
5184
|
Identification of a factor Xa-interactive site within residues 337-372 of the factor VIII heavy chain
|
|
5185
|
Identification of a factor Xa-interactive site within residues 337-372 of the factor VIII heavy chain
|
|
5186
|
Identification of a factor Xa-interactive site within residues 337-372 of the factor VIII heavy chain
|
|
5187
|
Identification of a factor Xa-interactive site within residues 337-372 of the factor VIII heavy chain
|
|
5188
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5189
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5190
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5191
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5192
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5193
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5194
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5195
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5196
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5197
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5198
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5199
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5200
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5201
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5202
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5203
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5204
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5205
|
Kinetic Properties of the glucose-6-phosphate and 6-phosphogluconate dehydrogenases from Corynebacterium ...
|
|
5227
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5228
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5229
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5230
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5231
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5232
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5233
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5234
|
Kinetic studies of rat liver hekokinase D('glucokinase') in non-co-operative conditions show an ordered ...
|
|
5235
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5236
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5237
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5238
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5239
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5240
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5241
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5242
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5243
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5244
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5245
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5246
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5247
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5248
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5249
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5250
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5251
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5252
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5253
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5254
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5255
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5256
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5257
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5258
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5259
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5260
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5261
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5262
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5263
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5264
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5265
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5266
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5267
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5268
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5269
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5270
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5271
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5272
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5273
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5274
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5275
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5276
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5277
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5278
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5279
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5280
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5281
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5282
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5283
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5284
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5285
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5286
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5287
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5288
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5289
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5290
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5291
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5292
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5293
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5294
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5295
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5296
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5297
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5298
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5299
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5300
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5301
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5302
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5303
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5304
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5305
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5306
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5307
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5308
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5309
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5310
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5311
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5312
|
Involvement of the chicken liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase sequence ...
|
|
5313
|
How does Pseudomonas fluorescens avoid suicide from its antibiotic pseudomonic acid?: Evidence for two ...
|
|
5314
|
How does Pseudomonas fluorescens avoid suicide from its antibiotic pseudomonic acid?: Evidence for two ...
|
|
5315
|
How does Pseudomonas fluorescens avoid suicide from its antibiotic pseudomonic acid?: Evidence for two ...
|
|
5316
|
Reaction mechanism of the heterotetrameric (alpha2beta2) E1 component of 2-oxo acid dehydrogenase multienzyme ...
|
|
5317
|
Reaction mechanism of the heterotetrameric (alpha2beta2) E1 component of 2-oxo acid dehydrogenase multienzyme ...
|
|
5318
|
Purification and characterization of rat skeletal muscle ...
|
|
5319
|
Purification and characterization of rat skeletal muscle ...
|
|
5320
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5321
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5322
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5323
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5324
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5325
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5326
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5327
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5328
|
Differences in kinetic properties of phospho and dephospho forms of fructose-6-phosphate, 2-kinase and ...
|
|
5329
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5330
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5331
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5332
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5333
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5334
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5335
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5336
|
L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12
|
|
5337
|
Structural, kinetic and calorimetric charecterization of the cold-active phosphoglycerate kinase from the ...
|
|
5338
|
Structural, kinetic and calorimetric charecterization of the cold-active phosphoglycerate kinase from the ...
|
|
5339
|
Structural, kinetic and calorimetric charecterization of the cold-active phosphoglycerate kinase from the ...
|
|
5340
|
Structural, kinetic and calorimetric charecterization of the cold-active phosphoglycerate kinase from the ...
|
|
5341
|
General base catalysis in the urate oxidase reaction: evidence for a novel Thr-Lys catalytic diad
|
|
5342
|
General base catalysis in the urate oxidase reaction: evidence for a novel Thr-Lys catalytic diad
|
|
5343
|
General base catalysis in the urate oxidase reaction: evidence for a novel Thr-Lys catalytic diad
|
|
5344
|
General base catalysis in the urate oxidase reaction: evidence for a novel Thr-Lys catalytic diad
|
|
5345
|
General base catalysis in the urate oxidase reaction: evidence for a novel Thr-Lys catalytic diad
|
|
5346
|
General base catalysis in the urate oxidase reaction: evidence for a novel Thr-Lys catalytic diad
|
|
5347
|
The catalytic role of the distal site asparagine-histidine couple in catalase-peroxidases
|
|
5348
|
The catalytic role of the distal site asparagine-histidine couple in catalase-peroxidases
|
|
5349
|
The catalytic role of the distal site asparagine-histidine couple in catalase-peroxidases
|
|
5350
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5351
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5352
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5353
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5354
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5355
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5356
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5357
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5358
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5359
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5360
|
Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose ...
|
|
5361
|
Vibrationally enhanced hydrogen tunneling in the Escherichia coli thymidylate synthase catalyzed reaction
|
|
5362
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5363
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5364
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5365
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5366
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5367
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5368
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5369
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5370
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5371
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5372
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5373
|
Sulfaphenazole derivatives as tools for comparing cytochrome P450 2C5 and human cytochromes P450 2Cs: ...
|
|
5374
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5375
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5376
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5377
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5378
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5379
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5380
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5381
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5382
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5383
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5384
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5385
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5386
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5387
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5388
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5389
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5390
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5391
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5392
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5393
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5394
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5395
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5396
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5397
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5398
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5399
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5400
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5401
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5402
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5403
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5404
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5405
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5406
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5407
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5408
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5409
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5410
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5411
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5412
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5413
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5414
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5415
|
Structural basis for the altered activity of Gly794 variants of Escherichia coli beta-galactosidase
|
|
5416
|
Positional isotope exchange analysis of the pantothenate synthetase reaction
|
|
5417
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5418
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5419
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5420
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5421
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5422
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5423
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5424
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5425
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5426
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5427
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5428
|
Mutation and evolution of the magnesium-binding site of a class II aminoacyl-tRNA synthetase
|
|
5429
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5430
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5431
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5432
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5433
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5434
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5435
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5436
|
Kinetic characterization of thiolate anion formation and chemical catalysis of activated microsomal ...
|
|
5437
|
S-methylmethionine is both a substrate and an inactivator of 1-aminocyclopropane-1-carboxylate synthase
|
|
5438
|
S-methylmethionine is both a substrate and an inactivator of 1-aminocyclopropane-1-carboxylate synthase
|
|
5439
|
Expression and regulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoenzymes white adipose ...
|
|
5440
|
Expression and regulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoenzymes white adipose ...
|
|
5441
|
Expression and regulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoenzymes white adipose ...
|
|
5442
|
Expression and regulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoenzymes white adipose ...
|
|
5443
|
Allosteric inhibition of PTP1B activity by selective modification of a non-active site cysteine residue
|
|
5444
|
Allosteric inhibition of PTP1B activity by selective modification of a non-active site cysteine residue
|
|
5445
|
Kinetic inactivation study of mushroom tyrosinase: intermediate detection by denaturants
|
|
5446
|
Kinetic inactivation study of mushroom tyrosinase: intermediate detection by denaturants
|
|
5447
|
Kinetic inactivation study of mushroom tyrosinase: intermediate detection by denaturants
|
|
5448
|
The deoxyfluoro-D-glucopyranose 6-phosphates and their effect on yeast glucose phosphate isomerase
|
|
5449
|
The deoxyfluoro-D-glucopyranose 6-phosphates and their effect on yeast glucose phosphate isomerase
|
|
5450
|
The deoxyfluoro-D-glucopyranose 6-phosphates and their effect on yeast glucose phosphate isomerase
|
|
5451
|
The deoxyfluoro-D-glucopyranose 6-phosphates and their effect on yeast glucose phosphate isomerase
|
|
5452
|
The deoxyfluoro-D-glucopyranose 6-phosphates and their effect on yeast glucose phosphate isomerase
|
|
5453
|
The deoxyfluoro-D-glucopyranose 6-phosphates and their effect on yeast glucose phosphate isomerase
|
|
5454
|
The deoxyfluoro-D-glucopyranose 6-phosphates and their effect on yeast glucose phosphate isomerase
|
|
5455
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5456
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5457
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5458
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5459
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5460
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5461
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5462
|
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5` to ...
|
|
5463
|
Reactivity study on microperoxidase-8
|
|
5464
|
Reactivity study on microperoxidase-8
|
|
5465
|
Oxidation of 2,4-dichlorophenol catalyzed by horseradish peroxidase: characterization of the reaction ...
|
|
5466
|
Oxidation of 2,4-dichlorophenol catalyzed by horseradish peroxidase: characterization of the reaction ...
|
|
5467
|
A variant of rabbit phosphoglucose isomerase
|
|
5468
|
A variant of rabbit phosphoglucose isomerase
|
|
5469
|
A variant of rabbit phosphoglucose isomerase
|
|
5470
|
A variant of rabbit phosphoglucose isomerase
|
|
5471
|
Astrocytic glucose-6-phosphatase and the permeability of brain microsomes to glucose 6-phosphate
|
|
5472
|
Astrocytic glucose-6-phosphatase and the permeability of brain microsomes to glucose 6-phosphate
|
|
5473
|
Astrocytic glucose-6-phosphatase and the permeability of brain microsomes to glucose 6-phosphate
|
|
5474
|
Astrocytic glucose-6-phosphatase and the permeability of brain microsomes to glucose 6-phosphate
|
|
5475
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5476
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5477
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5478
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5479
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5480
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5481
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5482
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5483
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5484
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5485
|
Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for ...
|
|
5486
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5487
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5488
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5489
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5490
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5491
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5492
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5493
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5494
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5495
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5496
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5497
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5498
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5499
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5500
|
Functions of the conserved anionic amino acids and those interacting with the substrate phosphate group of ...
|
|
5501
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5502
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5503
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5504
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5505
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5506
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5507
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5508
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5509
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5510
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5511
|
Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual ...
|
|
5512
|
pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase
|
|
5513
|
pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase
|
|
5514
|
pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase
|
|
5515
|
pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase
|
|
5516
|
pH and kinetic isotope effects on sarcosine oxidation by N-methyltryptophan oxidase
|
|
5517
|
Autocatalytic formation of green heme: evidence for H2O2-dependent formation of a covalent methionine-heme ...
|
|
5518
|
Autocatalytic formation of green heme: evidence for H2O2-dependent formation of a covalent methionine-heme ...
|
|
5519
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5520
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5521
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5522
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5523
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5524
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5525
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5526
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5527
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5528
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5529
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5530
|
Determination of the Affinity of Each Component of a Composite Quaternary Transition-State Analogue Complex of ...
|
|
5531
|
Purification and characterization of Go alpha and three types of Gi alpha after expression in Escherichia coli
|
|
5532
|
Purification and characterization of Go alpha and three types of Gi alpha after expression in Escherichia coli
|
|
5533
|
Purification and characterization of Go alpha and three types of Gi alpha after expression in Escherichia coli
|
|
5534
|
Purification and characterization of Go alpha and three types of Gi alpha after expression in Escherichia coli
|
|
5535
|
Production of the Neuromodulator H2S by Cystathionine beta-Synthase via the Condensation of Cysteine and ...
|
|
5536
|
Production of the Neuromodulator H2S by Cystathionine beta-Synthase via the Condensation of Cysteine and ...
|
|
5537
|
Acetylation of the chemotaxis response regulator CheY by acetyl-CoA synthetase purified from Escherichia coli
|
|
5538
|
The effect of pH and temperature on the kinetic parameters of phosphoglucose isomerase. Participation of ...
|
|
5539
|
The effect of pH and temperature on the kinetic parameters of phosphoglucose isomerase. Participation of ...
|
|
5540
|
The effect of pH and temperature on the kinetic parameters of phosphoglucose isomerase. Participation of ...
|
|
5541
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5542
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5543
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5544
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5545
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5546
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5547
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5548
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5549
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5550
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5551
|
Disruption of the aldolase A tetramer into catalytically active monomers
|
|
5552
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5553
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5554
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5555
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5556
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5557
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5558
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5559
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5560
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5561
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5562
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5563
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5564
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5565
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5566
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5567
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5568
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5569
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5570
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5571
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5572
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5573
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5574
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5575
|
The isozymes of glucose-phosphate isomerase (GPI-A2 and GPI-B2) from the teleost fish Fundulus heteroclitus ...
|
|
5576
|
Human ocular aldehyde dehydrogenase isozymes: distribution and properties as major soluble proteins in cornea ...
|
|
5577
|
Human ocular aldehyde dehydrogenase isozymes: distribution and properties as major soluble proteins in cornea ...
|
|
5578
|
Human ocular aldehyde dehydrogenase isozymes: distribution and properties as major soluble proteins in cornea ...
|
|
5579
|
Human ocular aldehyde dehydrogenase isozymes: distribution and properties as major soluble proteins in cornea ...
|
|
5580
|
Human ocular aldehyde dehydrogenase isozymes: distribution and properties as major soluble proteins in cornea ...
|
|
5581
|
Methadone metabolism by human placenta
|
|
5582
|
Methadone metabolism by human placenta
|
|
5583
|
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory ...
|
|
5584
|
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory ...
|
|
5585
|
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory ...
|
|
5586
|
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory ...
|
|
5587
|
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory ...
|
|
5588
|
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory ...
|
|
5589
|
A novel O-phospho-L-serine sulfhydrylation reaction catalyzed by O-acetylserine sulfhydrylase from Aeropyrum ...
|
|
5590
|
A novel O-phospho-L-serine sulfhydrylation reaction catalyzed by O-acetylserine sulfhydrylase from Aeropyrum ...
|
|
5591
|
A novel O-phospho-L-serine sulfhydrylation reaction catalyzed by O-acetylserine sulfhydrylase from Aeropyrum ...
|
|
5592
|
Kinetic mechanism of choline oxidase from Arthrobacter globiformis
|
|
5593
|
Kinetic mechanism of choline oxidase from Arthrobacter globiformis
|
|
5594
|
Kinetic mechanism of choline oxidase from Arthrobacter globiformis
|
|
5595
|
Kinetic mechanism of choline oxidase from Arthrobacter globiformis
|
|
5596
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5597
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5598
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5599
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5600
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5601
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5602
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5603
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5604
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5605
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5606
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5607
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5608
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5609
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5610
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5611
|
Phosphorylation of human erythrocyte pyruvate kinase by soluble cyclic-AMP-dependent protein kinases
|
|
5612
|
Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural ...
|
|
5613
|
Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural ...
|
|
5614
|
Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural ...
|
|
5615
|
Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural ...
|
|
5616
|
Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural ...
|
|
5617
|
Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural ...
|
|
5618
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5619
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5620
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5621
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5622
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5623
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5624
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5625
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5626
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5627
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5628
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5629
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5630
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5631
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5632
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5633
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5634
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5635
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5636
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5637
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5638
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5639
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5640
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5641
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5642
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5643
|
Kinetic modeling of beta-chloroprene metabolism: I. In vitro rates in liver and lung tissue fractions from ...
|
|
5644
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5645
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5646
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5647
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5648
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5649
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5650
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5651
|
Direct interaction between substrates and endogenous steroids in the active site may change the activity of ...
|
|
5652
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5653
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5654
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5655
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5656
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5657
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5658
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5659
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5660
|
D-Fucose metabolism in a pseudomonad. I. Oxidation of D-fucose to D-fucono- -lactone by a D-aldohexose ...
|
|
5661
|
Molecular dissection of human methionine synthase reductase: determination of the flavin redox potentials in ...
|
|
5662
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5663
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5664
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5665
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5666
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5667
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5668
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5669
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5670
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5671
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5672
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5673
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5674
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5675
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5676
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5677
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5678
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5679
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5680
|
Structural and regulatory properties of pyruvate kinase from the Cyanobacterium synechococcus PCC 6301
|
|
5681
|
Site-directed mutagenesis of active site residues of phosphite dehydrogenase
|
|
5682
|
Site-directed mutagenesis of active site residues of phosphite dehydrogenase
|
|
5683
|
Site-directed mutagenesis of active site residues of phosphite dehydrogenase
|
|
5684
|
Site-directed mutagenesis of active site residues of phosphite dehydrogenase
|
|
5685
|
Site-directed mutagenesis of active site residues of phosphite dehydrogenase
|
|
5686
|
Site-directed mutagenesis of active site residues of phosphite dehydrogenase
|
|
5687
|
Site-directed mutagenesis of active site residues of phosphite dehydrogenase
|
|
5688
|
Active Site Mutations of Pseudomonas aeruginosa Exotoxin A
|
|
5689
|
Active Site Mutations of Pseudomonas aeruginosa Exotoxin A
|
|
5690
|
Active Site Mutations of Pseudomonas aeruginosa Exotoxin A
|
|
5691
|
Active Site Mutations of Pseudomonas aeruginosa Exotoxin A
|
|
5692
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5693
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5694
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5695
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5696
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5697
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5698
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5699
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5700
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5701
|
Overexpression, purification, and characterization of VanX, a D-, D-dipeptidase which is essential for ...
|
|
5702
|
Identification of an archaeal alpha-L-fucosidase encoded by an interrupted gene. Production of a functional ...
|
|
5703
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5704
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5705
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5706
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5707
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5708
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5709
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5710
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5711
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5712
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5713
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5714
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5715
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5716
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5717
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5718
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5719
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5720
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5721
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5722
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5723
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5724
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5725
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5726
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5727
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5728
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5729
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5730
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5731
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5732
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5733
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5734
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5735
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5736
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5737
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5738
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5739
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5740
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5741
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5742
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5743
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5744
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5745
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5746
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5747
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5748
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5749
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5750
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5751
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5752
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5753
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5754
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5755
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5756
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5757
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5758
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5759
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5760
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5761
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5762
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5763
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5764
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5765
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5766
|
Kinetic analysis of phenylalanine dehydrogenase mutants designed for aliphatic amino acid dehydrogenase ...
|
|
5767
|
ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase
|
|
5768
|
ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase
|
|
5769
|
Human Acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty ...
|
|
5770
|
Human Acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty ...
|
|
5771
|
Human Acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty ...
|
|
5772
|
Kinetic mechanisms of glycine oxidase from Bacillus subtilis
|
|
5773
|
Kinetic mechanisms of glycine oxidase from Bacillus subtilis
|
|
5774
|
Kinetic mechanisms of glycine oxidase from Bacillus subtilis
|
|
5775
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5776
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5777
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5778
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5779
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5780
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5781
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5782
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5783
|
Steady-state and pre-steady-state kinetic analysis of halopropane conversion by a rhodococcus haloalkane ...
|
|
5784
|
Conformational flexibility of the C terminus with implications for substrate binding and catalysis revealed in ...
|
|
5785
|
Conformational flexibility of the C terminus with implications for substrate binding and catalysis revealed in ...
|
|
5786
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5787
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5788
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5789
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5790
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5791
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5792
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5793
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5794
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5795
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5796
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5797
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5798
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5799
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5800
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5801
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5802
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5803
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5804
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5805
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5806
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5807
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5808
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5809
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5810
|
Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from ...
|
|
5811
|
Characterization of recombinant xylitol dehydrogenase from Galactocandida mastotermitis expressed in ...
|
|
5812
|
Characterization of recombinant xylitol dehydrogenase from Galactocandida mastotermitis expressed in ...
|
|
5813
|
Characterization of recombinant xylitol dehydrogenase from Galactocandida mastotermitis expressed in ...
|
|
5814
|
Characterization of recombinant xylitol dehydrogenase from Galactocandida mastotermitis expressed in ...
|
|
5815
|
Characterization of recombinant xylitol dehydrogenase from Galactocandida mastotermitis expressed in ...
|
|
5816
|
Characterization of recombinant xylitol dehydrogenase from Galactocandida mastotermitis expressed in ...
|
|
5828
|
Agonist occupation of an alpha2A-adrenoreceptor-Gi1alpha fusion protein results in activation of both ...
|
|
5829
|
Agonist occupation of an alpha2A-adrenoreceptor-Gi1alpha fusion protein results in activation of both ...
|
|
5830
|
Characterization of a cytosolic malate dehydrogenase cDNA which encodes an isozyme toward oxaloacetate ...
|
|
5831
|
Characterization of a cytosolic malate dehydrogenase cDNA which encodes an isozyme toward oxaloacetate ...
|
|
5832
|
Characterization of a cytosolic malate dehydrogenase cDNA which encodes an isozyme toward oxaloacetate ...
|
|
5833
|
Characterization of a cytosolic malate dehydrogenase cDNA which encodes an isozyme toward oxaloacetate ...
|
|
5834
|
Isolation and characterization of tissue-specific isozymes of glucosephosphate isomerase from catfish and ...
|
|
5835
|
Isolation and characterization of tissue-specific isozymes of glucosephosphate isomerase from catfish and ...
|
|
5836
|
Isolation and characterization of tissue-specific isozymes of glucosephosphate isomerase from catfish and ...
|
|
5837
|
Isolation and characterization of tissue-specific isozymes of glucosephosphate isomerase from catfish and ...
|
|
5843
|
Evidence for heme-mediated redox regulation of human cystathionine beta-synthase activity
|
|
5844
|
Evidence for heme-mediated redox regulation of human cystathionine beta-synthase activity
|
|
5845
|
Glutathione Synthesis in Streptococcus agalactiae
|
|
5846
|
Glutathione Synthesis in Streptococcus agalactiae
|
|
5847
|
Glutathione Synthesis in Streptococcus agalactiae
|
|
5848
|
Glutathione Synthesis in Streptococcus agalactiae
|
|
5849
|
Glutathione Synthesis in Streptococcus agalactiae
|
|
5850
|
A single amino acid substitution (157 Gly----Val) in a phosphoglycerate kinase variant (PGK Shizuoka) ...
|
|
5851
|
A single amino acid substitution (157 Gly----Val) in a phosphoglycerate kinase variant (PGK Shizuoka) ...
|
|
5852
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5853
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5854
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5855
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5856
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5857
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5858
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5859
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5860
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5861
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5862
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5863
|
Kinetic mechanism and metabolic role of pyruvate phosphate dikinase from Entamoeba histolytica
|
|
5864
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5865
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5866
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5867
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5868
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5869
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5870
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5871
|
Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+ -specific isocitrate ...
|
|
5872
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5873
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5874
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5875
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5876
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5877
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5878
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5879
|
A mechanism of sulfite neurotoxicity direct inhibition of glutamate dehydrogenase
|
|
5880
|
Importance of product/reactant equilibration in the kinetics of the phosphoglucose isomerization reaction by ...
|
|
5881
|
Importance of product/reactant equilibration in the kinetics of the phosphoglucose isomerization reaction by ...
|
|
5882
|
Importance of product/reactant equilibration in the kinetics of the phosphoglucose isomerization reaction by ...
|
|
5883
|
Importance of product/reactant equilibration in the kinetics of the phosphoglucose isomerization reaction by ...
|
|
5884
|
Importance of product/reactant equilibration in the kinetics of the phosphoglucose isomerization reaction by ...
|
|
5885
|
Importance of product/reactant equilibration in the kinetics of the phosphoglucose isomerization reaction by ...
|
|
5886
|
Functional and molecular modelling studies of two hereditary fructose intolerance-causing mutations at ...
|
|
5887
|
Functional and molecular modelling studies of two hereditary fructose intolerance-causing mutations at ...
|
|
5888
|
Functional and molecular modelling studies of two hereditary fructose intolerance-causing mutations at ...
|
|
5889
|
Functional and molecular modelling studies of two hereditary fructose intolerance-causing mutations at ...
|
|
5890
|
Functional and molecular modelling studies of two hereditary fructose intolerance-causing mutations at ...
|
|
5891
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5892
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5893
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5894
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5895
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5896
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5897
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5898
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5899
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5900
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5901
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5902
|
The inhibition of acetate, pyruvate, and 3-phosphogylcerate kinases by chromium adenosine triphosphate
|
|
5903
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5904
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5905
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5906
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5907
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5908
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5909
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5910
|
Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA
|
|
5911
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5912
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5913
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5914
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5915
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5916
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5917
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5918
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5919
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5920
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5921
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5922
|
Properties and mechanism of human erythrocyte phosphoglycerate kinase
|
|
5923
|
Crystal structure of bacterial morphinone reductase and properties of the C191A mutant enzyme
|
|
5924
|
Crystal structure of bacterial morphinone reductase and properties of the C191A mutant enzyme
|
|
5925
|
Crystal structure of bacterial morphinone reductase and properties of the C191A mutant enzyme
|
|
5926
|
Crystal structure of bacterial morphinone reductase and properties of the C191A mutant enzyme
|
|
5927
|
Reaction of morphinone reductase with 2-cyclohexen-1-one and 1-nitrocyclohexene: proton donation, ligand ...
|
|
5928
|
Reaction of morphinone reductase with 2-cyclohexen-1-one and 1-nitrocyclohexene: proton donation, ligand ...
|
|
5929
|
Aldehyde dismutase activity of human liver alcohol dehydrogenase
|
|
5930
|
Aldehyde dismutase activity of human liver alcohol dehydrogenase
|
|
5931
|
Aldehyde dismutase activity of human liver alcohol dehydrogenase
|
|
5932
|
Correlated changes of some enzyme activities and cofactor and substrate contents of pea cotyledon tissue ...
|
|
5933
|
Correlated changes of some enzyme activities and cofactor and substrate contents of pea cotyledon tissue ...
|
|
5934
|
Correlated changes of some enzyme activities and cofactor and substrate contents of pea cotyledon tissue ...
|
|
5935
|
Catecholase activity associated with copper-S100B
|
|
5936
|
Catecholase activity associated with copper-S100B
|
|
5937
|
Catecholase activity associated with copper-S100B
|
|
5938
|
Catecholase activity associated with copper-S100B
|
|
5939
|
Catecholase activity associated with copper-S100B
|
|
5940
|
Catecholase activity associated with copper-S100B
|
|
5941
|
Catecholase activity associated with copper-S100B
|
|
5942
|
Catecholase activity associated with copper-S100B
|
|
5943
|
4-hydroxyretinoic acid, a novel substrate for human liver microsomal UDP-glucuronosyltransferase(s) and ...
|
|
5944
|
4-hydroxyretinoic acid, a novel substrate for human liver microsomal UDP-glucuronosyltransferase(s) and ...
|
|
5945
|
4-hydroxyretinoic acid, a novel substrate for human liver microsomal UDP-glucuronosyltransferase(s) and ...
|
|
5946
|
4-hydroxyretinoic acid, a novel substrate for human liver microsomal UDP-glucuronosyltransferase(s) and ...
|
|
5947
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5948
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5949
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5950
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5951
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5952
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5953
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5954
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5955
|
Influence of lipophilicity on the interactions of hydroxy stilbenes with cytochrome P450 3A4
|
|
5956
|
Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a ...
|
|
5957
|
Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a ...
|
|
5958
|
Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a ...
|
|
5959
|
Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a ...
|
|
5960
|
Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a ...
|
|
5961
|
Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a ...
|
|
5962
|
Alteration of substrate specificity by a naturally-occurring aldolase B mutation (Ala337-->Val) in fructose ...
|
|
5963
|
Alteration of substrate specificity by a naturally-occurring aldolase B mutation (Ala337-->Val) in fructose ...
|
|
5964
|
Alteration of substrate specificity by a naturally-occurring aldolase B mutation (Ala337-->Val) in fructose ...
|
|
5965
|
Alteration of substrate specificity by a naturally-occurring aldolase B mutation (Ala337-->Val) in fructose ...
|
|
5966
|
Partial purification, kinetic analysis, and amino acid sequence information of a flavonol ...
|
|
5967
|
Partial purification, kinetic analysis, and amino acid sequence information of a flavonol ...
|
|
5968
|
Partial purification, kinetic analysis, and amino acid sequence information of a flavonol ...
|
|
5969
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5970
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5971
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5972
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5973
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5974
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5975
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5976
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5977
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5978
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5979
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5980
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5981
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5982
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5983
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5984
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5985
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5986
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5987
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5988
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5989
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5990
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5991
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5992
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5993
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5994
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5995
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5996
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5997
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5998
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
5999
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|
|
6000
|
Identification of the enzymatic active site of CD38 by site-directed mutagenesis
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.