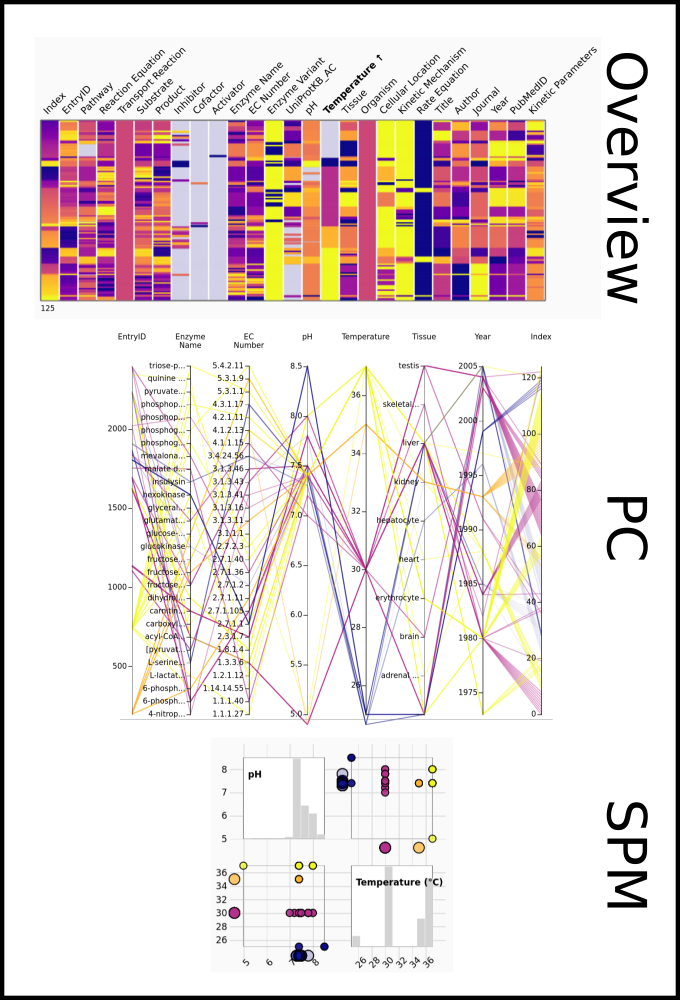
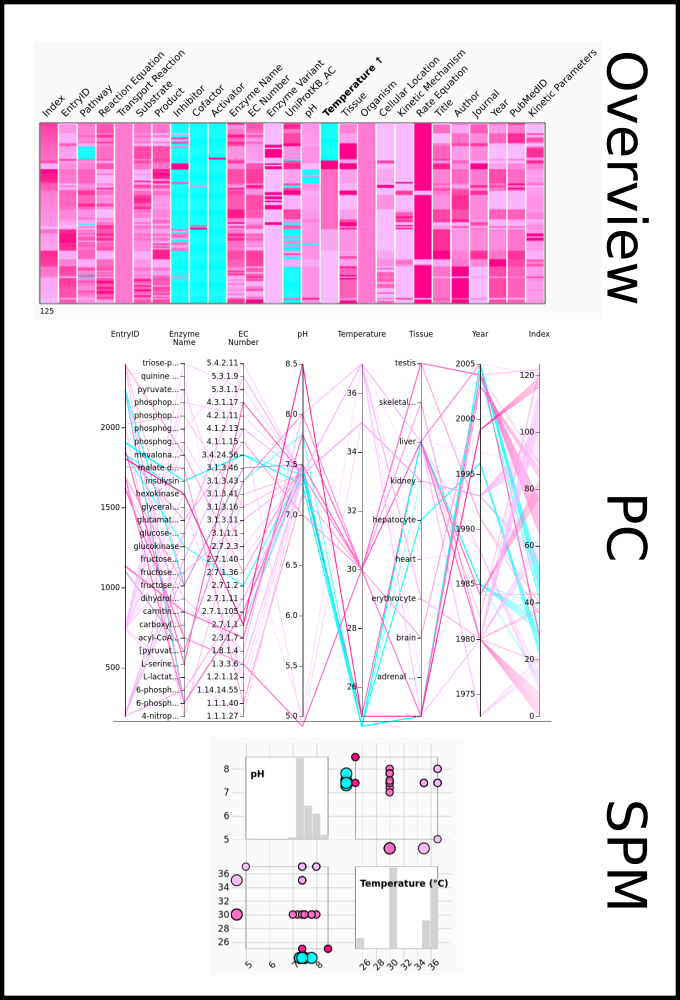
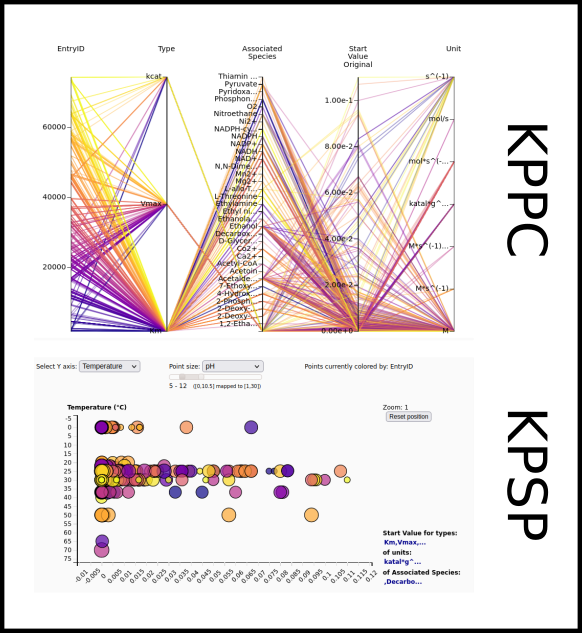
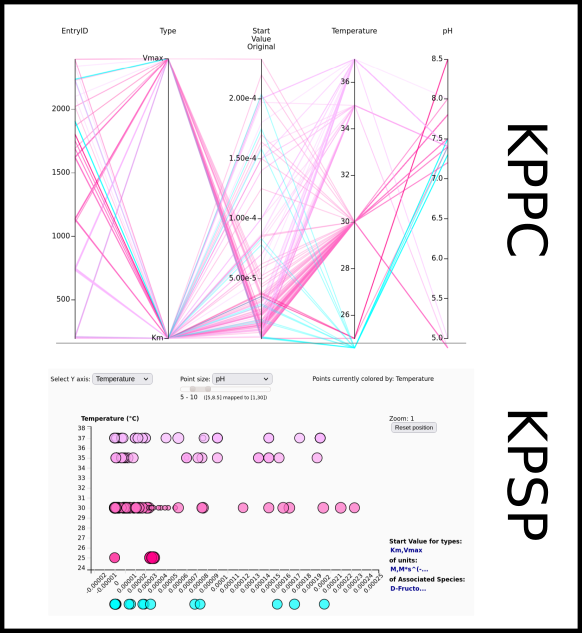
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
55001
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55002
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55003
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55004
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55005
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55006
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55007
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55008
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55009
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55010
|
Purification and characterization of soluble starch synthases from maize endosperm.
|
|
55011
|
Specificity of starch synthase isoforms from potato.
|
|
55012
|
Specificity of starch synthase isoforms from potato.
|
|
55013
|
Specificity of starch synthase isoforms from potato.
|
|
55014
|
Specificity of starch synthase isoforms from potato.
|
|
55015
|
Specificity of starch synthase isoforms from potato.
|
|
55016
|
Specificity of starch synthase isoforms from potato.
|
|
55017
|
Specificity of starch synthase isoforms from potato.
|
|
55018
|
Specificity of starch synthase isoforms from potato.
|
|
55019
|
Specificity of starch synthase isoforms from potato.
|
|
55020
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55021
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55022
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55023
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55024
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55025
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55026
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55027
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55028
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55029
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55030
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55031
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55032
|
Comparative studies of NADP-malic enzyme from C4- and C3-plants
|
|
55033
|
An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro ...
|
|
55034
|
An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro ...
|
|
55035
|
An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro ...
|
|
55036
|
An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro ...
|
|
55037
|
An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro ...
|
|
55038
|
An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro ...
|
|
55039
|
An increase in apparent affinity for sucrose of mung bean sucrose synthase is caused by in vitro ...
|
|
55040
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55041
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55042
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55043
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55044
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55045
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55046
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55047
|
A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells
|
|
55048
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55049
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55050
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55051
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55052
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55053
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55054
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55055
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55056
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55057
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55058
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55059
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55060
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55061
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55062
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55063
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55064
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55065
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55066
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55067
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55068
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55069
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55070
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55071
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55072
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55073
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55074
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55075
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55076
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55077
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55078
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55079
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55080
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55081
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55082
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55083
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55084
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55085
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55086
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55087
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55088
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55089
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55090
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55091
|
Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) ...
|
|
55092
|
Identification of the gene encoding homoserine kinase from Arabidopsis thaliana and characterization of the ...
|
|
55093
|
Purification and properties of phenylalanine ammonia-lyase from leaf mustard
|
|
55094
|
A simple procedure for purifying the major chloroplast fructose-1,6-bisphosphatase from spinach (Spinacia ...
|
|
55095
|
A simple procedure for purifying the major chloroplast fructose-1,6-bisphosphatase from spinach (Spinacia ...
|
|
55096
|
A simple procedure for purifying the major chloroplast fructose-1,6-bisphosphatase from spinach (Spinacia ...
|
|
55097
|
[3H]naloxone binding in the brain of the domestic chick (Gallus domesticus) determined by in vitro ...
|
|
55098
|
[3H]naloxone binding in the brain of the domestic chick (Gallus domesticus) determined by in vitro ...
|
|
55099
|
[3H]naloxone binding in the brain of the domestic chick (Gallus domesticus) determined by in vitro ...
|
|
55100
|
[3H]naloxone binding in the brain of the domestic chick (Gallus domesticus) determined by in vitro ...
|
|
55101
|
[3H]naloxone binding in the brain of the domestic chick (Gallus domesticus) determined by in vitro ...
|
|
55102
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55103
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55104
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55105
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55106
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55107
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55108
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55109
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55110
|
Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force
|
|
55111
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55112
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55113
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55114
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55115
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55116
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55117
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55118
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55119
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55120
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55121
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55122
|
Isolation and characterization of the cytosolic and chloroplastic 3-phosphoglycerate kinase from spinach ...
|
|
55123
|
Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate
|
|
55124
|
Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate
|
|
55125
|
Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate
|
|
55126
|
Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate
|
|
55127
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55128
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55129
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55130
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55131
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55132
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55133
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55134
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55135
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55136
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55137
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55138
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55139
|
On the interaction of substrate analogues with non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase ...
|
|
55140
|
Purification and partial characterization of pyruvate decarboxylase from Oryza sativa L.
|
|
55141
|
Purification and partial characterization of pyruvate decarboxylase from Oryza sativa L.
|
|
55142
|
Purification and partial characterization of pyruvate decarboxylase from Oryza sativa L.
|
|
55143
|
Purification and partial characterization of pyruvate decarboxylase from Oryza sativa L.
|
|
55144
|
Purification and partial characterization of pyruvate decarboxylase from Oryza sativa L.
|
|
55145
|
Partial Purification and Characterization of Complex I, NADH:Ubiquinone Reductase, from the Inner Membrane of ...
|
|
55146
|
Partial Purification and Characterization of Complex I, NADH:Ubiquinone Reductase, from the Inner Membrane of ...
|
|
55147
|
Recombinant expression, purification, and characterization of three isoenzymes of aspartate aminotransferase ...
|
|
55148
|
Recombinant expression, purification, and characterization of three isoenzymes of aspartate aminotransferase ...
|
|
55149
|
Recombinant expression, purification, and characterization of three isoenzymes of aspartate aminotransferase ...
|
|
55150
|
Recombinant expression, purification, and characterization of three isoenzymes of aspartate aminotransferase ...
|
|
55151
|
Recombinant expression, purification, and characterization of three isoenzymes of aspartate aminotransferase ...
|
|
55152
|
Recombinant expression, purification, and characterization of three isoenzymes of aspartate aminotransferase ...
|
|
55153
|
H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and ...
|
|
55154
|
H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and ...
|
|
55155
|
H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and ...
|
|
55156
|
H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and ...
|
|
55157
|
H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and ...
|
|
55158
|
H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and ...
|
|
55159
|
H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and ...
|
|
55160
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55161
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55162
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55163
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55164
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55165
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55166
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55167
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55168
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55169
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55170
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55171
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55172
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55173
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55174
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55175
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55176
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55177
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55178
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55179
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55180
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55181
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55182
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55183
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55184
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55185
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55186
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55187
|
Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
|
|
55188
|
Non-photosynthetic 'malic enzyme' from maize: a constituvely expressed enzyme that responds to plant defence ...
|
|
55189
|
Non-photosynthetic 'malic enzyme' from maize: a constituvely expressed enzyme that responds to plant defence ...
|
|
55190
|
Non-photosynthetic 'malic enzyme' from maize: a constituvely expressed enzyme that responds to plant defence ...
|
|
55191
|
Non-photosynthetic 'malic enzyme' from maize: a constituvely expressed enzyme that responds to plant defence ...
|
|
55192
|
Non-photosynthetic 'malic enzyme' from maize: a constituvely expressed enzyme that responds to plant defence ...
|
|
55193
|
Non-photosynthetic 'malic enzyme' from maize: a constituvely expressed enzyme that responds to plant defence ...
|
|
55194
|
Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the ...
|
|
55195
|
Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the ...
|
|
55196
|
Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the ...
|
|
55197
|
Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the ...
|
|
55198
|
Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the ...
|
|
55199
|
Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the ...
|
|
55200
|
Purification, characterization and immunological properties of 2,3-bisphosphoglycerate-independent ...
|
|
55201
|
Purification, characterization and immunological properties of 2,3-bisphosphoglycerate-independent ...
|
|
55202
|
Purification and characterization of the proton translocating plasma membrane ATPase of red beet storage ...
|
|
55203
|
Purification and characterization of the proton translocating plasma membrane ATPase of red beet storage ...
|
|
55204
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55205
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55206
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55207
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55208
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55209
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55210
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55211
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55212
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55213
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55214
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55215
|
Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional ...
|
|
55216
|
UDP-glucose pyrophosphorylase from potato tuber: purification and characterization.
|
|
55217
|
UDP-glucose pyrophosphorylase from potato tuber: purification and characterization.
|
|
55218
|
UDP-glucose pyrophosphorylase from potato tuber: purification and characterization.
|
|
55219
|
UDP-glucose pyrophosphorylase from potato tuber: purification and characterization.
|
|
55220
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55221
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55222
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55223
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55224
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55225
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55226
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55227
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55228
|
Pyrophosphorylases in Solanum tuberosum (IV. Purification, Tissue Localization, and Physicochemical Properties ...
|
|
55229
|
Purification and comparative characterization of an enolase from spinach.
|
|
55230
|
Purification and comparative characterization of an enolase from spinach.
|
|
55231
|
Purification and comparative characterization of an enolase from spinach.
|
|
55232
|
Purification and comparative characterization of an enolase from spinach.
|
|
55233
|
Purification and comparative characterization of an enolase from spinach.
|
|
55234
|
Purification and comparative characterization of an enolase from spinach.
|
|
55235
|
Purification and comparative characterization of an enolase from spinach.
|
|
55236
|
Purification and comparative characterization of an enolase from spinach.
|
|
55237
|
Purification and comparative characterization of an enolase from spinach.
|
|
55238
|
Purification and comparative characterization of an enolase from spinach.
|
|
55239
|
Purification and comparative characterization of an enolase from spinach.
|
|
55240
|
Purification and comparative characterization of an enolase from spinach.
|
|
55241
|
Purification and comparative characterization of an enolase from spinach.
|
|
55242
|
Purification and comparative characterization of an enolase from spinach.
|
|
55243
|
Regulation of photosynthetic carbon metabolism. The effect of inorganic phosphate on stromal ...
|
|
55244
|
Regulation of photosynthetic carbon metabolism. The effect of inorganic phosphate on stromal ...
|
|
55245
|
Regulation of photosynthetic carbon metabolism. The effect of inorganic phosphate on stromal ...
|
|
55246
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55247
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55248
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55249
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55250
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55251
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55252
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55253
|
The Role of Neuraminic Acid in the Stability and Enzymatic Activity of Acid Phosphatase of the Human Prostate ...
|
|
55254
|
Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia ...
|
|
55255
|
Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia ...
|
|
55256
|
Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia ...
|
|
55257
|
Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia ...
|
|
55258
|
Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia ...
|
|
55259
|
Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia ...
|
|
55260
|
Purification and properties of cystathionine beta-lyase from Arabidopsis thaliana overexpressed in Escherichia ...
|
|
55261
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55262
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55263
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55264
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55265
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55266
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55267
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55268
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55269
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55270
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55271
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55272
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55273
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55274
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55275
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55276
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55277
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55278
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55279
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55280
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55281
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55282
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55283
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55284
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55285
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55286
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55287
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55288
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55289
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55290
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55291
|
Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase.
|
|
55292
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55293
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55294
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55295
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55296
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55297
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55298
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55299
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55300
|
The catalytic site of chloroplastic NADP-dependent malate dehydrogenase contains a His/Asp pair.
|
|
55301
|
Kinetic properties of pure overproduced Bacillus subtilis phenylalanyl-tRNA synthetase do not favour its in ...
|
|
55302
|
Kinetic properties of pure overproduced Bacillus subtilis phenylalanyl-tRNA synthetase do not favour its in ...
|
|
55303
|
Kinetic properties of pure overproduced Bacillus subtilis phenylalanyl-tRNA synthetase do not favour its in ...
|
|
55304
|
Specific receptor for ceruloplasmin in membrane fragments from aortic and heart tissues
|
|
55305
|
Specific receptor for ceruloplasmin in membrane fragments from aortic and heart tissues
|
|
55306
|
Production and properties of recombinant C3-type phosphoenolpyruvate carboxylase from Sorghum vulgare: in ...
|
|
55307
|
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH ...
|
|
55308
|
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH ...
|
|
55309
|
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH ...
|
|
55310
|
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH ...
|
|
55311
|
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH ...
|
|
55312
|
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH ...
|
|
55313
|
Endothelin-converting enzyme-2 is a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH ...
|
|
55314
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55315
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55316
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55317
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55318
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55319
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55320
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55321
|
Comparison of the Kinetic Behavior toward Pyridine Nucleotides of NAD-Linked Dehydrogenases from Plant ...
|
|
55322
|
Sedoheptulose-1,7-bisphosphatase from wheat chloroplasts.
|
|
55323
|
Sedoheptulose-1,7-bisphosphatase from wheat chloroplasts.
|
|
55324
|
Sedoheptulose-1,7-bisphosphatase from wheat chloroplasts.
|
|
55325
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55326
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55327
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55328
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55329
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55330
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55331
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55332
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55333
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55334
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55335
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55336
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55337
|
Phenylalanine ammonia-lyase: a model for the cooperativity kinetics induced by D- and L-phenylalanine.
|
|
55338
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55339
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55340
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55341
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55342
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55343
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55344
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55345
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55346
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55347
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55348
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55349
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55350
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55351
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55352
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55353
|
Phenylalanine ammonia-lyase: mirror-image packing of D- and L-phenylalanine and D- and L-transition state ...
|
|
55354
|
Phenylalanine ammonia-lyase: purification and characterization from soybean cell suspension cultures.
|
|
55355
|
Phenylalanine ammonia-lyase: purification and characterization from soybean cell suspension cultures.
|
|
55356
|
Phenylalanine ammonia-lyase: purification and characterization from soybean cell suspension cultures.
|
|
55357
|
Phenylalanine ammonia-lyase: purification and characterization from soybean cell suspension cultures.
|
|
55358
|
Phenylalanine ammonia-lyase: purification and characterization from soybean cell suspension cultures.
|
|
55359
|
Transport of glycine, serine, and proline into spinach leaf mitochondria.
|
|
55360
|
Transport of glycine, serine, and proline into spinach leaf mitochondria.
|
|
55361
|
Transport of glycine, serine, and proline into spinach leaf mitochondria.
|
|
55362
|
Identification of glutamic acid 78 as the active site nucleophile in Bacillus subtilis xylanase using ...
|
|
55363
|
Identification of glutamic acid 78 as the active site nucleophile in Bacillus subtilis xylanase using ...
|
|
55364
|
Identification of glutamic acid 78 as the active site nucleophile in Bacillus subtilis xylanase using ...
|
|
55365
|
Identification of glutamic acid 78 as the active site nucleophile in Bacillus subtilis xylanase using ...
|
|
55366
|
Slow passive diffusion of NAD+ between intact isolated plant mitochondria and suspending medium.
|
|
55367
|
Slow passive diffusion of NAD+ between intact isolated plant mitochondria and suspending medium.
|
|
55368
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55369
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55370
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55371
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55372
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55373
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55374
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55375
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55376
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55377
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55378
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55379
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55380
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55381
|
Study of the arrangement of the functional domains along the yeast cytoplasmic aspartyl-tRNA synthetase
|
|
55382
|
NADPH-cytochrome P450 oxidoreductase from the chicken (Gallus gallus): sequence characterization, functional ...
|
|
55383
|
Proteolytic activity of bovine lactoferrin.
|
|
55387
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55388
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55389
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55390
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55391
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55392
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55393
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55394
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55395
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55396
|
The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves.
|
|
55397
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55398
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55399
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55400
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55401
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55402
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55403
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55404
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55405
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55406
|
Separation and characterization of four hexose kinases from developing maize kernels
|
|
55407
|
Conversion of aldonic acids to their corresponding 2-keto-3-deoxy-analogs by the non-carbohydrate enzyme ...
|
|
55408
|
Conversion of aldonic acids to their corresponding 2-keto-3-deoxy-analogs by the non-carbohydrate enzyme ...
|
|
55409
|
Conversion of aldonic acids to their corresponding 2-keto-3-deoxy-analogs by the non-carbohydrate enzyme ...
|
|
55410
|
ADP-glucose drives starch synthesis in isolated maize endosperm amyloplasts: characterization of starch ...
|
|
55411
|
Purification and characterization of N-acetylglutamate 5-phosphotransferase from pea (Pisum sativum) ...
|
|
55412
|
Purification and characterization of N-acetylglutamate 5-phosphotransferase from pea (Pisum sativum) ...
|
|
55413
|
Properties of Pyrophosphate:Fructose-6-Phosphate Phosphotransferase from Endosperm of Developing Wheat ...
|
|
55414
|
Properties of Pyrophosphate:Fructose-6-Phosphate Phosphotransferase from Endosperm of Developing Wheat ...
|
|
55415
|
Properties of Pyrophosphate:Fructose-6-Phosphate Phosphotransferase from Endosperm of Developing Wheat ...
|
|
55416
|
Properties of Pyrophosphate:Fructose-6-Phosphate Phosphotransferase from Endosperm of Developing Wheat ...
|
|
55417
|
Properties of Pyrophosphate:Fructose-6-Phosphate Phosphotransferase from Endosperm of Developing Wheat ...
|
|
55418
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55419
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55420
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55421
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55422
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55423
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55424
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55425
|
Light modulation of chloroplast membrane-bound ferredoxin-NADP+ oxidoreductase.
|
|
55426
|
A palmitoylation switch mechanism in the regulation of the visual cycle.
|
|
55427
|
A palmitoylation switch mechanism in the regulation of the visual cycle.
|
|
55428
|
A palmitoylation switch mechanism in the regulation of the visual cycle.
|
|
55429
|
Urinary lysosomal hydrolases in mucolipidosis II and mucolipidosis III
|
|
55430
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55431
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55432
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55433
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55434
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55435
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55436
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55437
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55438
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55439
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55440
|
Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic ...
|
|
55441
|
Structure and chromosomal localization of human and mouse genes for hematopoietic prostaglandin D synthase. ...
|
|
55442
|
Structure and chromosomal localization of human and mouse genes for hematopoietic prostaglandin D synthase. ...
|
|
55443
|
Structure and chromosomal localization of human and mouse genes for hematopoietic prostaglandin D synthase. ...
|
|
55444
|
Structure and chromosomal localization of human and mouse genes for hematopoietic prostaglandin D synthase. ...
|
|
55445
|
Biochemical and immunological characterization of rat spleen prostaglandin D synthetase
|
|
55446
|
Biochemical and immunological characterization of rat spleen prostaglandin D synthetase
|
|
55447
|
Biochemical and immunological characterization of rat spleen prostaglandin D synthetase
|
|
55448
|
Biochemical and immunological characterization of rat spleen prostaglandin D synthetase
|
|
55449
|
Biochemical and immunological characterization of rat spleen prostaglandin D synthetase
|
|
55450
|
Galactokinase of Vicia faba seeds.
|
|
55451
|
Galactokinase of Vicia faba seeds.
|
|
55452
|
Galactokinase of Vicia faba seeds.
|
|
55453
|
Galactokinase of Vicia faba seeds.
|
|
55454
|
Galactokinase of Vicia faba seeds.
|
|
55455
|
Galactokinase of Vicia faba seeds.
|
|
55456
|
Galactokinase of Vicia faba seeds.
|
|
55457
|
Isolation of inositol 1,3,4-trisphosphate 5/6-kinase, cDNA cloning and expression of the recombinant enzyme.
|
|
55458
|
Solubilization and reconstitution of chick renal mitochondrial 25-hydroxyvitamin D3 24-hydroxylase.
|
|
55459
|
Solubilization and reconstitution of kidney 25-hydroxyvitamin D3 1 alpha- and 24-hydroxylases from vitamin ...
|
|
55460
|
Serine hydroxymethyltransferase from soybean root nodules : purification and kinetic properties.
|
|
55461
|
Serine hydroxymethyltransferase from soybean root nodules : purification and kinetic properties.
|
|
55462
|
Serine hydroxymethyltransferase from soybean root nodules : purification and kinetic properties.
|
|
55463
|
Serine hydroxymethyltransferase from soybean root nodules : purification and kinetic properties.
|
|
55464
|
Serine hydroxymethyltransferase from soybean root nodules : purification and kinetic properties.
|
|
55465
|
Serine hydroxymethyltransferase from soybean root nodules : purification and kinetic properties.
|
|
55466
|
Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) ...
|
|
55467
|
Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) ...
|
|
55468
|
Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) ...
|
|
55469
|
Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) ...
|
|
55470
|
Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) ...
|
|
55471
|
Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) ...
|
|
55472
|
Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate.
|
|
55473
|
Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate.
|
|
55474
|
Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate.
|
|
55475
|
Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate.
|
|
55476
|
Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate.
|
|
55477
|
Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate.
|
|
55478
|
Inhibition of ribose-5-phosphate isomerase by 4-phosphoerythronate.
|
|
55479
|
Expression and purification of human mPGES-1 in E. coli and identification of inhibitory compounds from a ...
|
|
55480
|
Expression and purification of human mPGES-1 in E. coli and identification of inhibitory compounds from a ...
|
|
55481
|
Expression and purification of human mPGES-1 in E. coli and identification of inhibitory compounds from a ...
|
|
55482
|
Characterization of dimethylargininase from bovine brain: evidence for a zinc binding site.
|
|
55483
|
Characterization of dimethylargininase from bovine brain: evidence for a zinc binding site.
|
|
55484
|
Characterization of dimethylargininase from bovine brain: evidence for a zinc binding site.
|
|
55485
|
Characterization of dimethylargininase from bovine brain: evidence for a zinc binding site.
|
|
55486
|
Biochemical and immunological studies of purified mouse beta-galactosidase
|
|
55487
|
Biochemical and immunological studies of purified mouse beta-galactosidase
|
|
55488
|
Biochemical and immunological studies of purified mouse beta-galactosidase
|
|
55489
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55490
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55491
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55492
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55493
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55494
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55495
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55496
|
The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
|
|
55497
|
Cloning and recombinant expression of rat and human kynureninase.
|
|
55498
|
Cloning and recombinant expression of rat and human kynureninase.
|
|
55499
|
Cloning and recombinant expression of rat and human kynureninase.
|
|
55500
|
Cloning and recombinant expression of rat and human kynureninase.
|
|
55501
|
Cloning and recombinant expression of rat and human kynureninase.
|
|
55502
|
Cloning and recombinant expression of rat and human kynureninase.
|
|
55503
|
Cloning and recombinant expression of rat and human kynureninase.
|
|
55504
|
2-Keto-4-hydroxyglutarate aldolase: purification and characterization of the homogeneous enzyme from bovine ...
|
|
55505
|
2-Keto-4-hydroxyglutarate aldolase: purification and characterization of the homogeneous enzyme from bovine ...
|
|
55506
|
2-Keto-4-hydroxyglutarate aldolase: purification and characterization of the homogeneous enzyme from bovine ...
|
|
55507
|
2-Keto-4-hydroxyglutarate aldolase: purification and characterization of the homogeneous enzyme from bovine ...
|
|
55508
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55509
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55510
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55511
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55512
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55513
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55514
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55515
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55516
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55517
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55518
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55519
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55520
|
Excision of products of oxidative DNA base damage by human NTH1 protein.
|
|
55521
|
Molecular cloning and functional characterization of a rainbow trout liver Oatp
|
|
55522
|
Molecular cloning and functional characterization of a rainbow trout liver Oatp
|
|
55523
|
Molecular cloning and functional characterization of a rainbow trout liver Oatp
|
|
55524
|
Cysteine metabolism in cultured tobacco cells.
|
|
55525
|
The purification and characterization of ADP-glucose pyrophosphorylase A from developing maize seeds.
|
|
55526
|
The purification and characterization of ADP-glucose pyrophosphorylase A from developing maize seeds.
|
|
55527
|
Cold lability of pyruvate, orthophosphate dikinase in the maize leaf.
|
|
55528
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55529
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55530
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55531
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55532
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55533
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55534
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55535
|
Properties of Freshly Purified and Thiol-Treated Spinach Chloroplast Fructose Bisphosphatase
|
|
55536
|
Purification and biochemical characterization of some of the properties of recombinant human kynureninase.
|
|
55537
|
Purification and biochemical characterization of some of the properties of recombinant human kynureninase.
|
|
55538
|
Purification and biochemical characterization of some of the properties of recombinant human kynureninase.
|
|
55539
|
Purification and biochemical characterization of some of the properties of recombinant human kynureninase.
|
|
55540
|
Purification and biochemical characterization of some of the properties of recombinant human kynureninase.
|
|
55541
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55542
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55543
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55544
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55545
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55546
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55547
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55548
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55549
|
Tissue localization of barley (Hordeum vulgare) glutamine synthetase isoenzymes.
|
|
55550
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55551
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55552
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55553
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55554
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55555
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55556
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55557
|
Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of ...
|
|
55558
|
Purification and characterization of hydroxypyruvate reductase from a serine-producing methylotroph, ...
|
|
55559
|
Purification and characterization of hydroxypyruvate reductase from a serine-producing methylotroph, ...
|
|
55560
|
Purification and characterization of hydroxypyruvate reductase from a serine-producing methylotroph, ...
|
|
55561
|
Purification and characterization of hydroxypyruvate reductase from a serine-producing methylotroph, ...
|
|
55562
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55563
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55564
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55565
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55566
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55567
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55568
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55569
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55570
|
Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of ...
|
|
55571
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55572
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55573
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55574
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55575
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55576
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55577
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55578
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55579
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55580
|
Phosphorylation of serine residues 3, 6, 10, and 13 distinguishes membrane anchored from soluble glutamic acid ...
|
|
55581
|
Interaction of NADPH and triazine dyes with ferredoxin-NADP+ oxidoreductase.
|
|
55582
|
Interaction of NADPH and triazine dyes with ferredoxin-NADP+ oxidoreductase.
|
|
55583
|
Interaction of NADPH and triazine dyes with ferredoxin-NADP+ oxidoreductase.
|
|
55584
|
In Mycoplasma hominis the OppA-mediated cytoadhesion depends on its ATPase activity
|
|
55585
|
In Mycoplasma hominis the OppA-mediated cytoadhesion depends on its ATPase activity
|
|
55586
|
In Mycoplasma hominis the OppA-mediated cytoadhesion depends on its ATPase activity
|
|
55587
|
In Mycoplasma hominis the OppA-mediated cytoadhesion depends on its ATPase activity
|
|
55588
|
In Mycoplasma hominis the OppA-mediated cytoadhesion depends on its ATPase activity
|
|
55589
|
Glycine decarboxylase multienzyme complex. Purification and partial characterization from pea leaf ...
|
|
55590
|
Molecular cloning, functional characterization, and subcellular localization of soybean nodule ...
|
|
55591
|
Molecular cloning, functional characterization, and subcellular localization of soybean nodule ...
|
|
55592
|
Molecular cloning, functional characterization, and subcellular localization of soybean nodule ...
|
|
55593
|
Separation and allosteric properties of two forms of UDP-glucuronate carboxy-lyase.
|
|
55594
|
Separation and allosteric properties of two forms of UDP-glucuronate carboxy-lyase.
|
|
55595
|
Separation and allosteric properties of two forms of UDP-glucuronate carboxy-lyase.
|
|
55596
|
Separation and allosteric properties of two forms of UDP-glucuronate carboxy-lyase.
|
|
55597
|
Separation and allosteric properties of two forms of UDP-glucuronate carboxy-lyase.
|
|
55598
|
Separation and allosteric properties of two forms of UDP-glucuronate carboxy-lyase.
|
|
55599
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55600
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55601
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55602
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55603
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55604
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55605
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55606
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55607
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55608
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55609
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55610
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55611
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55612
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55613
|
Kinetic and structural characterization of spinach carbonic anhydrase.
|
|
55614
|
Cloning, expression and functional characterization of cytochrome P450 3A37 from turkey liver with high ...
|
|
55615
|
Cloning, expression and functional characterization of cytochrome P450 3A37 from turkey liver with high ...
|
|
55616
|
Cloning, expression and functional characterization of cytochrome P450 3A37 from turkey liver with high ...
|
|
55617
|
Cloning, expression and functional characterization of cytochrome P450 3A37 from turkey liver with high ...
|
|
55618
|
Cloning, expression and functional characterization of cytochrome P450 3A37 from turkey liver with high ...
|
|
55619
|
Aspartate aminotransferase from Panicum maximum Jacq. var. trichoglume Eyles, a C4 plant: purification, ...
|
|
55620
|
Aspartate aminotransferase from Panicum maximum Jacq. var. trichoglume Eyles, a C4 plant: purification, ...
|
|
55621
|
Limited proteolysis of inactive tetrameric chloroplast NADP-malate dehydrogenase produces active dimers.
|
|
55622
|
Limited proteolysis of inactive tetrameric chloroplast NADP-malate dehydrogenase produces active dimers.
|
|
55623
|
Phenylalanine ammonia-lyase from cotton (Gossypium hirsutum) hypocotyls: properties of the enzyme induced by a ...
|
|
55624
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55625
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55626
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55627
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55628
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55629
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55630
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55631
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55632
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55633
|
In vitro cytochrome P450-mediated hepatic activities for five substrates in specific pathogen free chickens.
|
|
55634
|
Enzymes for synthesis of 10-formyltetrahydrofolate in plants. Characterization of a monofunctional ...
|
|
55635
|
Enzymes for synthesis of 10-formyltetrahydrofolate in plants. Characterization of a monofunctional ...
|
|
55636
|
Enzymes for synthesis of 10-formyltetrahydrofolate in plants. Characterization of a monofunctional ...
|
|
55637
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55638
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55639
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55640
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55641
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55642
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55643
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55644
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55645
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55646
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55647
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55648
|
Characterization of chicken cytochrome P450 1A4 and 1A5: inter-paralog comparisons of substrate preference and ...
|
|
55649
|
A neuraminidase from Streptococcus sanguis that can release O-acetylated sialic acids
|
|
55650
|
A neuraminidase from Streptococcus sanguis that can release O-acetylated sialic acids
|
|
55651
|
Soluble forms of gamma-glutamyltransferase in human adult liver, fetal liver, and primary hepatoma compared
|
|
55652
|
Soluble forms of gamma-glutamyltransferase in human adult liver, fetal liver, and primary hepatoma compared
|
|
55653
|
Soluble forms of gamma-glutamyltransferase in human adult liver, fetal liver, and primary hepatoma compared
|
|
55654
|
Soluble forms of gamma-glutamyltransferase in human adult liver, fetal liver, and primary hepatoma compared
|
|
55655
|
Soluble forms of gamma-glutamyltransferase in human adult liver, fetal liver, and primary hepatoma compared
|
|
55656
|
Soluble forms of gamma-glutamyltransferase in human adult liver, fetal liver, and primary hepatoma compared
|
|
55657
|
IGF-I binding in primary culture of muscle cells of rainbow trout: changes during in vitro development
|
|
55658
|
IGF-I binding in primary culture of muscle cells of rainbow trout: changes during in vitro development
|
|
55659
|
IGF-I binding in primary culture of muscle cells of rainbow trout: changes during in vitro development
|
|
55660
|
IGF-I binding in primary culture of muscle cells of rainbow trout: changes during in vitro development
|
|
55661
|
IGF-I binding in primary culture of muscle cells of rainbow trout: changes during in vitro development
|
|
55662
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55663
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55664
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55665
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55666
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55667
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55668
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55669
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55670
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55671
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55672
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55673
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55674
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55675
|
Enzymatic properties of the beta-galactoside alpha 1 leads to 2 fucosyltransferase from porcine submaxillary ...
|
|
55676
|
Oxygen supply from the bird's eye perspective: globin E is a respiratory protein in the chicken retina.
|
|
55677
|
Oxygen supply from the bird's eye perspective: globin E is a respiratory protein in the chicken retina.
|
|
55678
|
Oxygen supply from the bird's eye perspective: globin E is a respiratory protein in the chicken retina.
|
|
55679
|
Oxygen supply from the bird's eye perspective: globin E is a respiratory protein in the chicken retina.
|
|
55680
|
Comparison of the effect of acidic inhibitors upon anaerobic phosphate uptake and dinitrophenol extrusion by ...
|
|
55681
|
Comparison of the effect of acidic inhibitors upon anaerobic phosphate uptake and dinitrophenol extrusion by ...
|
|
55682
|
Comparison of the effect of acidic inhibitors upon anaerobic phosphate uptake and dinitrophenol extrusion by ...
|
|
55683
|
Comparison of the effect of acidic inhibitors upon anaerobic phosphate uptake and dinitrophenol extrusion by ...
|
|
55684
|
Comparison of the effect of acidic inhibitors upon anaerobic phosphate uptake and dinitrophenol extrusion by ...
|
|
55685
|
Comparison of the effect of acidic inhibitors upon anaerobic phosphate uptake and dinitrophenol extrusion by ...
|
|
55686
|
Succinate oxidase in Neurospora.
|
|
55687
|
Succinate oxidase in Neurospora.
|
|
55688
|
Succinate oxidase in Neurospora.
|
|
55689
|
Succinate oxidase in Neurospora.
|
|
55690
|
Succinate oxidase in Neurospora.
|
|
55691
|
Succinate oxidase in Neurospora.
|
|
55692
|
Succinate oxidase in Neurospora.
|
|
55693
|
Succinate oxidase in Neurospora.
|
|
55694
|
Studies of human kidney gamma-glutamyl transpeptidase. Purification and structural, kinetic and immunological ...
|
|
55695
|
Partial purification and some properties of gamma-glutamyl transpeptidase from human bile.
|
|
55696
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55697
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55698
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55699
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55700
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55701
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55702
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55703
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55704
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55705
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55706
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55707
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55708
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55709
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55710
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55711
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55712
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55713
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55714
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55715
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55716
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55717
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55718
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55719
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55720
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55721
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55722
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55723
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55724
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55725
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55726
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55727
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55728
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55729
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55730
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55731
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55732
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55733
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55734
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55735
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55736
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55737
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55738
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55739
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55740
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55741
|
In vitro biotransformation and investigation of metabolic enzymes possibly responsible for the metabolism of ...
|
|
55742
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55743
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55744
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55745
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55746
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55747
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55748
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55749
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55750
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55751
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55752
|
Insensitivity of barley endosperm ADP-glucose pyrophosphorylase to 3-phosphoglycerate and orthophosphate ...
|
|
55753
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55754
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55755
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55756
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55757
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55758
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55759
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55760
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55761
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55762
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55763
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55764
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55765
|
Is leaf ADP-glucose pyrophosphorylase an allosteric enzyme?
|
|
55766
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55767
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55768
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55769
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55770
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55771
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55772
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55773
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55774
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55775
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55776
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55777
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55778
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55779
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55780
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55781
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55782
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55783
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55784
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55785
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55786
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55787
|
Kinetic mechanism and regulation of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves.
|
|
55788
|
Conserved amino acids near the carboxy terminus of bacterial tyrosyl-tRNA synthetase are involved in tRNA and ...
|
|
55789
|
Conserved amino acids near the carboxy terminus of bacterial tyrosyl-tRNA synthetase are involved in tRNA and ...
|
|
55790
|
Conserved amino acids near the carboxy terminus of bacterial tyrosyl-tRNA synthetase are involved in tRNA and ...
|
|
55791
|
Conserved amino acids near the carboxy terminus of bacterial tyrosyl-tRNA synthetase are involved in tRNA and ...
|
|
55792
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55793
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55794
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55795
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55796
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55797
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55798
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55799
|
Site-directed mutagenesis of rabbit LAT1 at amino acids 219 and 234.
|
|
55800
|
Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone ...
|
|
55801
|
Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone ...
|
|
55802
|
Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone ...
|
|
55803
|
Evidence that tetrahydrofolate does not bind to serine hydroxymethyltransferase with positive homotropic ...
|
|
55804
|
Human placental sialidase: partial purification and characterization
|
|
55805
|
Human placental sialidase: partial purification and characterization
|
|
55806
|
Human placental sialidase: partial purification and characterization
|
|
55807
|
Human placental sialidase: partial purification and characterization
|
|
55808
|
Human placental sialidase: partial purification and characterization
|
|
55809
|
Human placental sialidase: partial purification and characterization
|
|
55810
|
Human placental sialidase: partial purification and characterization
|
|
55811
|
Nudix hydrolase controls nucleotides and glycolytic mechanisms in hypoxic Aspergillus nidulans
|
|
55812
|
Nudix hydrolase controls nucleotides and glycolytic mechanisms in hypoxic Aspergillus nidulans
|
|
55813
|
Nudix hydrolase controls nucleotides and glycolytic mechanisms in hypoxic Aspergillus nidulans
|
|
55814
|
Nudix hydrolase controls nucleotides and glycolytic mechanisms in hypoxic Aspergillus nidulans
|
|
55815
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55816
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55817
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55818
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55819
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55820
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55821
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55822
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55823
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55824
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55825
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55826
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55827
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55828
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55829
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55830
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55831
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55832
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55833
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55834
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55835
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55836
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55837
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55838
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55839
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55840
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55841
|
The participation of human hepatic P450 isoforms, flavin-containing monooxygenases and aldehyde oxidase in the ...
|
|
55842
|
Enzymatic characterization and inhibitor discovery of a new cystathionine {gamma}-synthase from Helicobacter ...
|
|
55843
|
Enzymatic characterization and inhibitor discovery of a new cystathionine {gamma}-synthase from Helicobacter ...
|
|
55844
|
Enzymatic characterization and inhibitor discovery of a new cystathionine {gamma}-synthase from Helicobacter ...
|
|
55845
|
Enzymatic characterization and inhibitor discovery of a new cystathionine {gamma}-synthase from Helicobacter ...
|
|
55846
|
Enzymatic characterization and inhibitor discovery of a new cystathionine {gamma}-synthase from Helicobacter ...
|
|
55847
|
Enzymatic characterization and inhibitor discovery of a new cystathionine {gamma}-synthase from Helicobacter ...
|
|
55848
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55849
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55850
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55851
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55852
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55853
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55854
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55855
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55856
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55857
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55858
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55859
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55860
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55861
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55862
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55863
|
Cloning and characterization of a bifunctional leukotriene A(4) hydrolase from Saccharomyces cerevisiae.
|
|
55864
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55865
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55866
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55867
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55868
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55869
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55870
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55871
|
Characterization and separation of the arachidonic acid 5-lipoxygenase and linoleic acid omega-6 lipoxygenase ...
|
|
55872
|
Purification and properties of spinach leaf cytoplasmic fructose-1,6-bisphosphatase.
|
|
55873
|
Purification and properties of spinach leaf cytoplasmic fructose-1,6-bisphosphatase.
|
|
55874
|
Purification and properties of spinach leaf cytoplasmic fructose-1,6-bisphosphatase.
|
|
55875
|
Kinetic behavior of 25-hydroxyvitamin D-1-hydroxylase and -24-hydroxylase in rat kidney mitochondria.
|
|
55876
|
Kinetic behavior of 25-hydroxyvitamin D-1-hydroxylase and -24-hydroxylase in rat kidney mitochondria.
|
|
55877
|
Kinetic behavior of 25-hydroxyvitamin D-1-hydroxylase and -24-hydroxylase in rat kidney mitochondria.
|
|
55878
|
Kinetic behavior of 25-hydroxyvitamin D-1-hydroxylase and -24-hydroxylase in rat kidney mitochondria.
|
|
55879
|
25-Hydroxyvitamin D3-24-hydroxylase in rat kidney mitochondria.
|
|
55880
|
25-Hydroxyvitamin D3-24-hydroxylase in rat kidney mitochondria.
|
|
55881
|
An epimerase-reductase in L-fucose synthesis.
|
|
55882
|
An epimerase-reductase in L-fucose synthesis.
|
|
55883
|
Neuraminidase production by a Streptococcus sanguis strain associated with subacute bacterial endocarditis
|
|
55884
|
Glycosyltransferase activities and the differentiation of human promyelocytic (HL60) cells by retinoic acid ...
|
|
55885
|
Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and ...
|
|
55886
|
Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and ...
|
|
55887
|
Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and ...
|
|
55888
|
Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and ...
|
|
55889
|
Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and ...
|
|
55890
|
Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and ...
|
|
55891
|
Duodenase, a new serine protease of unusual specificity from bovine duodenal mucosa. Purification and ...
|
|
55892
|
Properties and regulation of Mg2+-dependent chloroplast inorganic pyrophosphatase from Sorghum vulgare leaves.
|
|
55893
|
Properties and regulation of Mg2+-dependent chloroplast inorganic pyrophosphatase from Sorghum vulgare leaves.
|
|
55894
|
Wheat invertases : characterization of cell wall-bound and soluble forms.
|
|
55895
|
Wheat invertases : characterization of cell wall-bound and soluble forms.
|
|
55896
|
Wheat invertases : characterization of cell wall-bound and soluble forms.
|
|
55897
|
Pyruvate Decarboxylase from Zea mays L. : I. Purification and Partial Characterization from Mature Kernels and ...
|
|
55898
|
Pyruvate Decarboxylase from Zea mays L. : I. Purification and Partial Characterization from Mature Kernels and ...
|
|
55899
|
Pyruvate Decarboxylase from Zea mays L. : I. Purification and Partial Characterization from Mature Kernels and ...
|
|
55900
|
Pyruvate Decarboxylase from Zea mays L. : I. Purification and Partial Characterization from Mature Kernels and ...
|
|
55901
|
Pyruvate Decarboxylase from Zea mays L. : I. Purification and Partial Characterization from Mature Kernels and ...
|
|
55902
|
Pyruvate Decarboxylase from Zea mays L. : I. Purification and Partial Characterization from Mature Kernels and ...
|
|
55903
|
Pyruvate Decarboxylase from Zea mays L. : I. Purification and Partial Characterization from Mature Kernels and ...
|
|
55904
|
Identification and functional characterization of a Na+-independent large neutral amino acid transporter, ...
|
|
55905
|
Identification and functional characterization of a Na+-independent large neutral amino acid transporter, ...
|
|
55906
|
Cytosolic NADP phosphatases I and II from Arthrobacter sp. strain KM: implication in regulation of NAD+/NADP+ ...
|
|
55907
|
Cytosolic NADP phosphatases I and II from Arthrobacter sp. strain KM: implication in regulation of NAD+/NADP+ ...
|
|
55908
|
Cytosolic NADP phosphatases I and II from Arthrobacter sp. strain KM: implication in regulation of NAD+/NADP+ ...
|
|
55909
|
Cytosolic NADP phosphatases I and II from Arthrobacter sp. strain KM: implication in regulation of NAD+/NADP+ ...
|
|
55910
|
Characterization of human liver (4-mythylumbelliferyl-alpha-D-N-acetylneuraminic acid) neuraminidase activity
|
|
55911
|
Human alpha-fucosidase. Single residual enzymatic form in fucosidosis
|
|
55912
|
Human alpha-fucosidase. Single residual enzymatic form in fucosidosis
|
|
55913
|
Human alpha-fucosidase. Single residual enzymatic form in fucosidosis
|
|
55914
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55915
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55916
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55917
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55918
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55919
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55920
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55921
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55922
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55923
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55924
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55925
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55926
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55927
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55928
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55929
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55930
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55931
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55932
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55933
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55934
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55935
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55936
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55937
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55938
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55939
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55940
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55941
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55942
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55943
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55944
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55945
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55946
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55947
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55948
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55949
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55950
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55951
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55952
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55953
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55954
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55955
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55956
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55957
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55958
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55959
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55960
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55961
|
A new regulatory principle for in vivo biochemistry: pleiotropic low affinity regulation by the adenine ...
|
|
55962
|
Purification and characterization of monomeric lysine decarboxylase from soybean (Glycine max) axes
|
|
55963
|
Purification and characterization of monomeric lysine decarboxylase from soybean (Glycine max) axes
|
|
55964
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55965
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55966
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55967
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55968
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55969
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55970
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55971
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55972
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55973
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55974
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55975
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55976
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55977
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55978
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55979
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55980
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55981
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55982
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55983
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55984
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55985
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55986
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55987
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55988
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55989
|
Purification and regulatory properties of mung bean (vigna radiata L.) serine hydroxymethyltransferase
|
|
55990
|
Improved purification of pyruvate decarboxylase from wheat germ. Its partial characterisation and comparison ...
|
|
55991
|
Improved purification of pyruvate decarboxylase from wheat germ. Its partial characterisation and comparison ...
|
|
55992
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
55993
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
55994
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
55995
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
55996
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
55997
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
55998
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
55999
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|
|
56000
|
Changing Kinetic Properties of Fructose-1,6-bisphosphatase from Pea Chloroplasts during Photosynthetic ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.