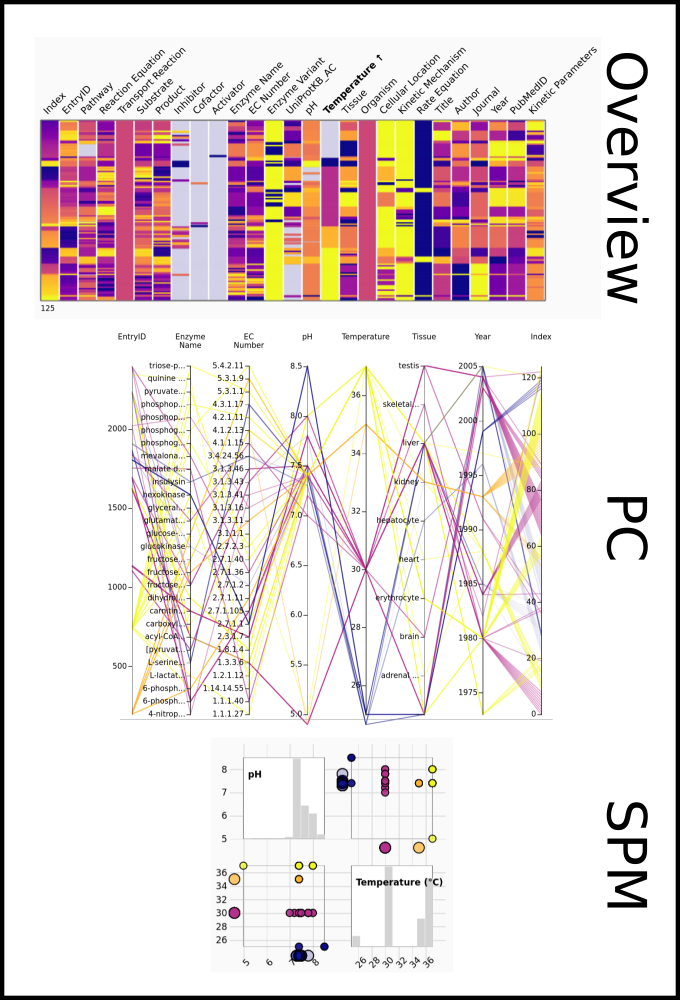
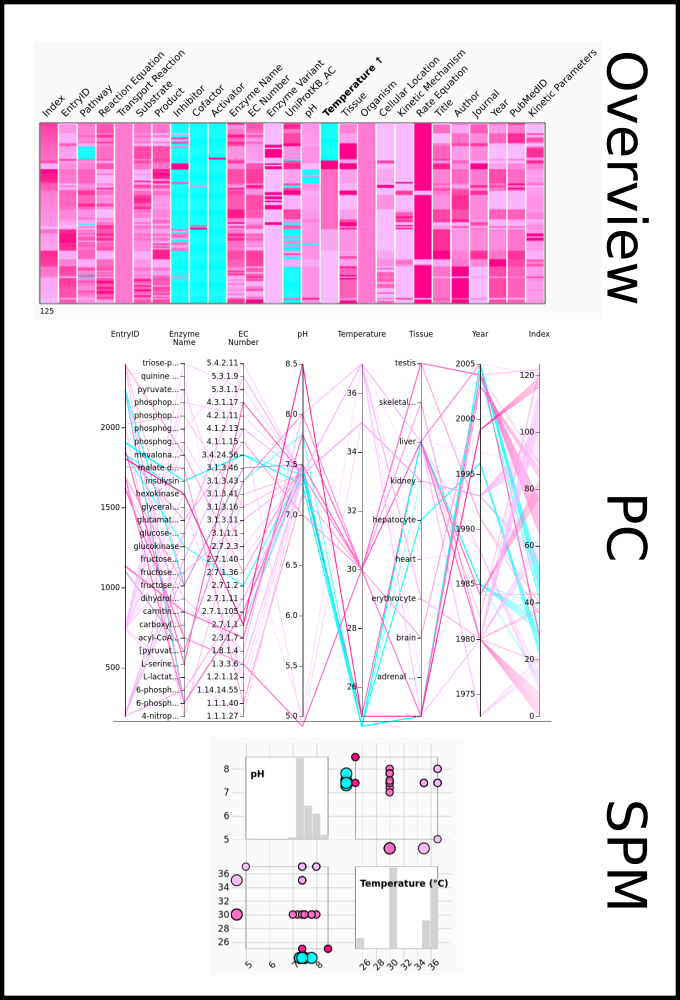
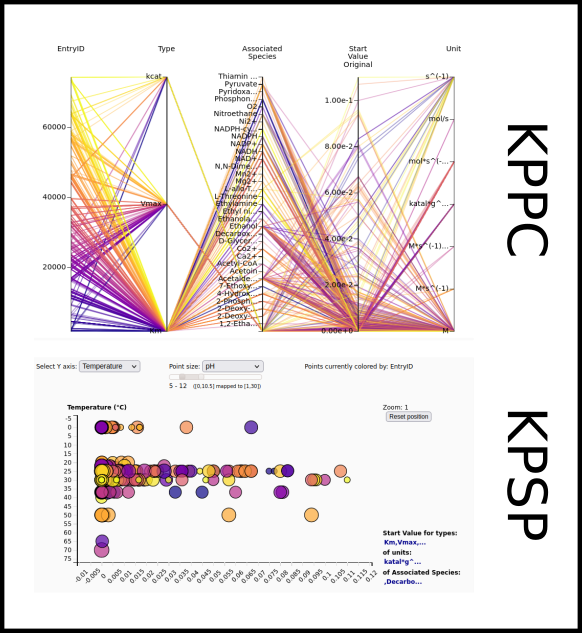
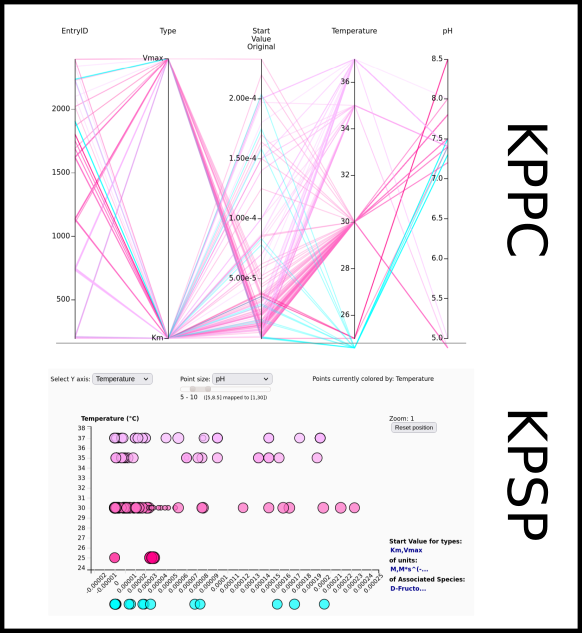
To improve SABIO-RK data interoperability, semantic markup was added to web pages as described and defined by
This structured information makes it easier to discover, collate and analyse our data.
|
9001
|
An antibody specific for coagulation factor IX enhances the activity of the intrinsic factor X-activating ...
|
|
9002
|
An antibody specific for coagulation factor IX enhances the activity of the intrinsic factor X-activating ...
|
|
9003
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9004
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9005
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9006
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9007
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9008
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9009
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9010
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9011
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9012
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9013
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9014
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9015
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9016
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9017
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9018
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9019
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9020
|
The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and ...
|
|
9021
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9022
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9023
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9024
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9025
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9026
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9027
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9028
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9029
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9030
|
Directed Mutations in the poorly defined region of porcine liver fructose-1,6-biphosphatase significantly ...
|
|
9031
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9032
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9033
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9034
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9035
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9036
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9037
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9038
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9039
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9040
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9041
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9042
|
Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of ...
|
|
9043
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9044
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9045
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9046
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9047
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9048
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9049
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9050
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9051
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9052
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9053
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9054
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9055
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9056
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9057
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9058
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9059
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9060
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9061
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9062
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9063
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9064
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9065
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9066
|
The proton transfer step catalyzed by yeast pyruvate kinase.
|
|
9067
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9068
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9069
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9070
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9071
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9072
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9073
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9074
|
Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in ...
|
|
9075
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9076
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9077
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9078
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9079
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9080
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9081
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9082
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9083
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9084
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9085
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9086
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9087
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9088
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9089
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9090
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9091
|
Kinetic characterization, structure modelling studies and crystallization of Trypanosoma brucei enolase
|
|
9092
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9093
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9094
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9095
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9096
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9097
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9098
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9099
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9100
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9101
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9102
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9103
|
The Phosphonopyruvate decarboxylase from Bacteroides fragilis
|
|
9104
|
Mechanistic Studies of a Flavin-Dependent Thymidylate Synthase
|
|
9105
|
Crystal structure of a novel shikimate dehydrogenase from Haemophilus influenzae
|
|
9106
|
Crystal structure of a novel shikimate dehydrogenase from Haemophilus influenzae
|
|
9107
|
Crystal structure of a novel shikimate dehydrogenase from Haemophilus influenzae
|
|
9108
|
The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a ...
|
|
9109
|
The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a ...
|
|
9110
|
The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a ...
|
|
9111
|
The catalytic role of glutamate 151 in the leucine aminopeptidase from Aeromonas proteolytica
|
|
9112
|
The catalytic role of glutamate 151 in the leucine aminopeptidase from Aeromonas proteolytica
|
|
9113
|
The catalytic role of glutamate 151 in the leucine aminopeptidase from Aeromonas proteolytica
|
|
9114
|
Interaction between D-glyceraldehyde-3-phosphate dehydrogenase and 3-phosphoglycerate kinase and its ...
|
|
9115
|
Interaction between D-glyceraldehyde-3-phosphate dehydrogenase and 3-phosphoglycerate kinase and its ...
|
|
9116
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9117
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9118
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9119
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9120
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9121
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9122
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9123
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9124
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9125
|
Purification and properties of ornithine carbamoyltransferase from loggerhead turtle liver
|
|
9126
|
Ornithine carbamoyl transferase activity
|
|
9127
|
Ornithine carbamoyl transferase activity
|
|
9128
|
Ornithine carbamoyl transferase activity
|
|
9129
|
Ornithine carbamoyl transferase activity
|
|
9130
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9131
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9132
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9133
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9134
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9135
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9136
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9137
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9138
|
Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate ...
|
|
9139
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9140
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9141
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9142
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9143
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9144
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9145
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9146
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9147
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9148
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9149
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9150
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9151
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9152
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9153
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9154
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9155
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9156
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9157
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9158
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9159
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9160
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9161
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9162
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9163
|
Active Site Mutants of Escherichia coli Dethiobiotin Synthetase: Effects of Mutations on Enzyme Catalytic and ...
|
|
9164
|
Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: insights into the ...
|
|
9165
|
Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: insights into the ...
|
|
9166
|
Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: insights into the ...
|
|
9167
|
Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: insights into the ...
|
|
9168
|
Cloning, expression, and characterization of a cis-3-chloroacrylic acid dehalogenase: insights into the ...
|
|
9169
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9170
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9171
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9172
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9173
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9174
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9175
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9176
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9177
|
New type of hexameric ornithine carbamoyltransferase with arginase activity in the cephamycin producers ...
|
|
9178
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9179
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9180
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9181
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9182
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9183
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9184
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9185
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9186
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9187
|
Active Site Mutants of Escherichia coli Citrate Synthase. Effects of Mutations on Catalytic and Allosteric ...
|
|
9188
|
In Vitro Mutagenesis of Escherichia coli Citrate Synthase to Clarify the Locations of Ligand Binding Sites
|
|
9189
|
In Vitro Mutagenesis of Escherichia coli Citrate Synthase to Clarify the Locations of Ligand Binding Sites
|
|
9190
|
In Vitro Mutagenesis of Escherichia coli Citrate Synthase to Clarify the Locations of Ligand Binding Sites
|
|
9191
|
In Vitro Mutagenesis of Escherichia coli Citrate Synthase to Clarify the Locations of Ligand Binding Sites
|
|
9192
|
In Vitro Mutagenesis of Escherichia coli Citrate Synthase to Clarify the Locations of Ligand Binding Sites
|
|
9193
|
In Vitro Mutagenesis of Escherichia coli Citrate Synthase to Clarify the Locations of Ligand Binding Sites
|
|
9194
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9195
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9196
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9197
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9198
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9199
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9200
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9201
|
Purification and characterization of lactate dehydrogenase isoenzymes 1 and 2 from Molinema dessetae ...
|
|
9202
|
Potent growth inhibition of human tumor cells in culture by arginine deiminase purified from a culture medium ...
|
|
9203
|
Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using ...
|
|
9204
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9205
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9206
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9207
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9208
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9209
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9210
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9211
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9212
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9213
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9214
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9215
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9216
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9217
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9218
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9219
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9220
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9221
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9222
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9223
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9224
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9225
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9226
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9227
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9228
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9229
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9230
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9231
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9232
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9233
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9234
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9235
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9236
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9237
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9238
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9239
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9240
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9241
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9242
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9243
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9244
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9245
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9246
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9247
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9248
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9249
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9250
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9251
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9252
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9253
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9254
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9255
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9256
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9257
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9258
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9259
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9260
|
Nature of the rat brain 6-Phosphofructo-1-kinase isozymes
|
|
9261
|
Expanding tRNA recognition of a tRNA synthetase by a single amino acid change
|
|
9262
|
Expanding tRNA recognition of a tRNA synthetase by a single amino acid change
|
|
9263
|
Expanding tRNA recognition of a tRNA synthetase by a single amino acid change
|
|
9264
|
Expanding tRNA recognition of a tRNA synthetase by a single amino acid change
|
|
9265
|
Expanding tRNA recognition of a tRNA synthetase by a single amino acid change
|
|
9266
|
Expanding tRNA recognition of a tRNA synthetase by a single amino acid change
|
|
9267
|
Expanding tRNA recognition of a tRNA synthetase by a single amino acid change
|
|
9268
|
Arginase of bacillus brevis nagano: purification, properties, and implication in gramicidin S biosynthesis
|
|
9269
|
Arginase of bacillus brevis nagano: purification, properties, and implication in gramicidin S biosynthesis
|
|
9270
|
Arginase of bacillus brevis nagano: purification, properties, and implication in gramicidin S biosynthesis
|
|
9271
|
Kinetics of glucosephosphate isomerase activity on fructose in the presence of arsenate. Reactivity of ...
|
|
9272
|
Kinetics of glucosephosphate isomerase activity on fructose in the presence of arsenate. Reactivity of ...
|
|
9273
|
Kinetics of glucosephosphate isomerase activity on fructose in the presence of arsenate. Reactivity of ...
|
|
9274
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9275
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9276
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9277
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9278
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9279
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9280
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9281
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9282
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9283
|
Modification of glyceraldehyde-3-phosphate dehydrogenase, EC 1.2.1.12, isolated from rainbow trout during ...
|
|
9284
|
Dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the ...
|
|
9285
|
Dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the ...
|
|
9286
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9287
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9288
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9289
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9290
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9291
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9292
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9293
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9294
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9295
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9296
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9297
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9298
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9299
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9300
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9301
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9302
|
Stereoselectivity of interaction of phosphoenolpyruvate analogues with various phosphoenolpyruvate-utilizing ...
|
|
9303
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9304
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9305
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9306
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9307
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9308
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9309
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9310
|
cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity
|
|
9311
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9312
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9313
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9314
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9315
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9316
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9317
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9318
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9319
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9320
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9321
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9322
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9323
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9324
|
Nucleotide specificity of Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase Kinetics, fluorescence ...
|
|
9332
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9333
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9334
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9335
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9336
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9337
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9338
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9339
|
Influence of compartmental localization on the function of yeast NADP+-specific isocitrate dehydrogenases
|
|
9340
|
IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the beta-oxidation of ...
|
|
9341
|
IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the beta-oxidation of ...
|
|
9342
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9343
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9344
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9345
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9346
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9347
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9348
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9349
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9350
|
The inhibition of yeast enolase by Li+ and Na+1
|
|
9351
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9352
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9353
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9354
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9355
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9356
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9357
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9358
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9359
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9360
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9361
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9362
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9363
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9364
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9365
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9366
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9367
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9368
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9369
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9370
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9371
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9372
|
Conversion of Methylamine Dehydrogenase to a Long-Chain Amine Dehydrogenase by Mutagenesis of a Single Residue
|
|
9373
|
The position of a key tyrosine in dTDP-4-Keto-6-deoxy-D-glucose-5-epimerase (EvaD) alters the substrate ...
|
|
9374
|
The position of a key tyrosine in dTDP-4-Keto-6-deoxy-D-glucose-5-epimerase (EvaD) alters the substrate ...
|
|
9375
|
The position of a key tyrosine in dTDP-4-Keto-6-deoxy-D-glucose-5-epimerase (EvaD) alters the substrate ...
|
|
9376
|
The position of a key tyrosine in dTDP-4-Keto-6-deoxy-D-glucose-5-epimerase (EvaD) alters the substrate ...
|
|
9377
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9378
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9379
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9380
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9381
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9382
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9383
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9384
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9385
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9386
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9387
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9388
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9389
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9390
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9391
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9392
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9393
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9394
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9395
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9396
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9397
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9398
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9399
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9400
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9401
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9402
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9403
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9404
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9405
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9406
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9407
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9408
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9409
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9410
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9411
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9412
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9413
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9414
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9415
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9416
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9417
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9418
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9419
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9420
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9421
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9422
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9423
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9424
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9425
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9426
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9427
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9428
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9429
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9430
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9431
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9432
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9433
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9434
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9435
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9436
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9437
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9438
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9439
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9440
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9441
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9442
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9443
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9444
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9445
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9446
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9447
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9448
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9449
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9450
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9451
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9452
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9453
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9454
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9455
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9456
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9457
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9458
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9459
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9460
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9461
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9462
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9463
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9464
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9465
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9466
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9467
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9468
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9469
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9470
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9471
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9472
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9473
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9474
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9475
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9476
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9477
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9478
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9479
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9480
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9481
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9482
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9483
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9484
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9485
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9486
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9487
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9488
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9489
|
Acetylcholinesterase Peripheral Anionic Site Degeneracy Conferred by Amino Acid Arrays Sharing a Common Core
|
|
9490
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9491
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9492
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9493
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9494
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9495
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9496
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9497
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9498
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9499
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9500
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9501
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9502
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9503
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9504
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9505
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9506
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9507
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9508
|
Kinetic and Site-directed Mutagenesis Studies of the Cysteine Residues of Bovine Low Molecular Weight ...
|
|
9509
|
A non-classical type of alveolar macrophage response to Trichinella spiralis infection
|
|
9510
|
A non-classical type of alveolar macrophage response to Trichinella spiralis infection
|
|
9511
|
A non-classical type of alveolar macrophage response to Trichinella spiralis infection
|
|
9512
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9513
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9514
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9515
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9516
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9517
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9518
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9519
|
Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase
|
|
9520
|
Mechanistic studies on phosphopantothenoylcysteine decarboxylase: trapping of an enethiolate intermediate with ...
|
|
9521
|
Mechanistic studies on phosphopantothenoylcysteine decarboxylase: trapping of an enethiolate intermediate with ...
|
|
9522
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9523
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9524
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9525
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9526
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9527
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9528
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9529
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9530
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9531
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9532
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9533
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9534
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9535
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9536
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9537
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9538
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9539
|
Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form ...
|
|
9540
|
Yeast phosphoglycerate kinase: investigation of catalytic function by site-directed mutagenesis
|
|
9541
|
Yeast phosphoglycerate kinase: investigation of catalytic function by site-directed mutagenesis
|
|
9542
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9543
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9544
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9545
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9546
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9547
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9548
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9549
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9550
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9551
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9552
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9553
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9554
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9555
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9556
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9557
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9558
|
The Role of Cysteine 78 in Fluorosulfonylbenzoyladenosine Inactivation of Rat Liver S-Adenosylhomocysteine ...
|
|
9559
|
Comparison of biochemical properties of liver arginase from streptozocin-induced diabetic and control mice
|
|
9560
|
Comparison of biochemical properties of liver arginase from streptozocin-induced diabetic and control mice
|
|
9561
|
Comparison of biochemical properties of liver arginase from streptozocin-induced diabetic and control mice
|
|
9562
|
Comparison of biochemical properties of liver arginase from streptozocin-induced diabetic and control mice
|
|
9563
|
Comparison of biochemical properties of liver arginase from streptozocin-induced diabetic and control mice
|
|
9564
|
Comparison of biochemical properties of liver arginase from streptozocin-induced diabetic and control mice
|
|
9565
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9566
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9567
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9568
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9569
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9570
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9571
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9572
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9573
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9574
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9575
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9576
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9577
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9578
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9579
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9580
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9581
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9582
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9583
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9584
|
The allosteric regulation of Pyruvate kinase.A site-directed mutagenesis study
|
|
9585
|
Redesigning the quaternary structure of R67 dihydrofolate reductase. Creation of an active monomer from a ...
|
|
9586
|
Redesigning the quaternary structure of R67 dihydrofolate reductase. Creation of an active monomer from a ...
|
|
9587
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9588
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9589
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9590
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9591
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9592
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9593
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9594
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9595
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9596
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9597
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9598
|
Role of Lys-32 residues in R67 dihydrofolate reductase probed by asymmetric mutations
|
|
9599
|
Influence of the unusual covalent adduct on the kinetics and formation of radical intermediates in ...
|
|
9600
|
Influence of the unusual covalent adduct on the kinetics and formation of radical intermediates in ...
|
|
9601
|
Influence of the unusual covalent adduct on the kinetics and formation of radical intermediates in ...
|
|
9602
|
Influence of the unusual covalent adduct on the kinetics and formation of radical intermediates in ...
|
|
9603
|
Isolation and Properties of Cytoplasmic alpha-Glycerol 3-Phosphate Dehydrogenase from the Pectoral Muscle of ...
|
|
9604
|
Isolation and Properties of Cytoplasmic alpha-Glycerol 3-Phosphate Dehydrogenase from the Pectoral Muscle of ...
|
|
9605
|
Isolation and Properties of Cytoplasmic alpha-Glycerol 3-Phosphate Dehydrogenase from the Pectoral Muscle of ...
|
|
9606
|
Isolation and Properties of Cytoplasmic alpha-Glycerol 3-Phosphate Dehydrogenase from the Pectoral Muscle of ...
|
|
9607
|
Isolation and Properties of Cytoplasmic alpha-Glycerol 3-Phosphate Dehydrogenase from the Pectoral Muscle of ...
|
|
9608
|
Cysteine-scanning mutagenesis of muscle carnitine palmitoyltransferase I reveals a single cysteine residue ...
|
|
9609
|
Cysteine-scanning mutagenesis of muscle carnitine palmitoyltransferase I reveals a single cysteine residue ...
|
|
9610
|
Cysteine-scanning mutagenesis of muscle carnitine palmitoyltransferase I reveals a single cysteine residue ...
|
|
9611
|
Cysteine-scanning mutagenesis of muscle carnitine palmitoyltransferase I reveals a single cysteine residue ...
|
|
9612
|
Role of the Proximal Ligand in Peroxidase Catalysis. Crystallographic, Kinetic, and Spectral Studies of ...
|
|
9613
|
Role of the Proximal Ligand in Peroxidase Catalysis. Crystallographic, Kinetic, and Spectral Studies of ...
|
|
9614
|
Role of the Proximal Ligand in Peroxidase Catalysis. Crystallographic, Kinetic, and Spectral Studies of ...
|
|
9615
|
Role of the Proximal Ligand in Peroxidase Catalysis. Crystallographic, Kinetic, and Spectral Studies of ...
|
|
9616
|
Role of the Proximal Ligand in Peroxidase Catalysis. Crystallographic, Kinetic, and Spectral Studies of ...
|
|
9617
|
Role of the Proximal Ligand in Peroxidase Catalysis. Crystallographic, Kinetic, and Spectral Studies of ...
|
|
9618
|
Role of the Proximal Ligand in Peroxidase Catalysis. Crystallographic, Kinetic, and Spectral Studies of ...
|
|
9619
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9620
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9621
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9622
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9623
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9624
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9625
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9626
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9627
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9628
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9629
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9630
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9631
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9632
|
Site-directed mutagenesis of yeast phosphoglycerate kinase. Arginines 65, 121 and 168
|
|
9633
|
Total conversion of bifunctional catalase-peroxidase (KatG) to monofunctional peroxidase by exchange of a ...
|
|
9634
|
Total conversion of bifunctional catalase-peroxidase (KatG) to monofunctional peroxidase by exchange of a ...
|
|
9635
|
Substrate binding to phosphoglycerate kinase monitored by 1-anilino-8-naphthalenesulfonate
|
|
9636
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9637
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9638
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9639
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9640
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9641
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9642
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9643
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9644
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9645
|
(E)-3-Cyanophosphoenolpyruvate, a new inhibitor of phosphoenolpyruvate-dependent enzymes
|
|
9646
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9647
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9648
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9649
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9650
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9651
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9652
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9653
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9654
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9655
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9656
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9657
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9658
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9659
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9660
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9661
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9662
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9663
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9664
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9665
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9666
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9667
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9668
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9669
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9670
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9671
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9672
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9673
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9674
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9675
|
Site-directed mutagenesis of the active site region in the Quinate/Shikimate 5-Dehydrogenase YdiB of ...
|
|
9676
|
Inhibition of apple polyphenol oxidase activity by procyanidins and polyphenol oxidation products
|
|
9677
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9678
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9679
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9680
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9681
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9682
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9683
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9684
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9685
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9686
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9687
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9688
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9689
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9690
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9691
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9692
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9693
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9694
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9695
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9696
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9697
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9698
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9699
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9700
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9701
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9702
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9703
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9704
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9705
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9706
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9707
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9708
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9709
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9710
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9711
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9712
|
Metal binding Asp-120 in metallo-beta-lactamase L1 from Stenotrophomonas maltophilia plays a crucial role in ...
|
|
9713
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9714
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9715
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9716
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9717
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9718
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9719
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9720
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9721
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9722
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9723
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9724
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9725
|
Substrate inhibition of acetylcholinesterase: residues affecting signal transduction from the surface to the ...
|
|
9726
|
Isolation of buffalo muscle aldolase and comparison of its properties with those of rabbit muscle aldolase
|
|
9727
|
Isolation of buffalo muscle aldolase and comparison of its properties with those of rabbit muscle aldolase
|
|
9728
|
Molecular cloning, expression, and properties of an unusual aldo-keto reductase family enzyme, pyridoxal ...
|
|
9729
|
Molecular cloning, expression, and properties of an unusual aldo-keto reductase family enzyme, pyridoxal ...
|
|
9730
|
Molecular cloning, expression, and properties of an unusual aldo-keto reductase family enzyme, pyridoxal ...
|
|
9731
|
Molecular cloning, expression, and properties of an unusual aldo-keto reductase family enzyme, pyridoxal ...
|
|
9732
|
Molecular cloning, expression, and properties of an unusual aldo-keto reductase family enzyme, pyridoxal ...
|
|
9733
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9734
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9735
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9736
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9737
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9738
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9739
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9740
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9741
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9742
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9743
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9744
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9745
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9746
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9747
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9748
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9749
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9750
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9751
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9752
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9753
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9754
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9755
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9756
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9757
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9758
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9759
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9760
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9761
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9762
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9763
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9764
|
Pyranose 2-oxidase from Phanerochaete chrysosporium--further biochemical characterisation
|
|
9765
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9766
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9767
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9768
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9769
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9770
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9771
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9772
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9773
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9774
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9775
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9776
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9777
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9778
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9779
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9780
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9781
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9782
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9783
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9784
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9785
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9786
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9787
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9788
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9789
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9790
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9791
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9792
|
Critical residues for the coenzyme specificity of NAD+-dependent 15-hydroxyprostaglandin dehydrogenase
|
|
9793
|
Biosynthesis of colitose: expression, purification, and mechanistic characterization of ...
|
|
9794
|
Biosynthesis of colitose: expression, purification, and mechanistic characterization of ...
|
|
9795
|
Biosynthesis of colitose: expression, purification, and mechanistic characterization of ...
|
|
9796
|
Creatine kinase in serum: 1. Determination of optimum reaction conditions
|
|
9797
|
Creatine kinase in serum: 1. Determination of optimum reaction conditions
|
|
9798
|
Creatine kinase in serum: 1. Determination of optimum reaction conditions
|
|
9799
|
Creatine kinase in serum: 1. Determination of optimum reaction conditions
|
|
9800
|
Identification of a human valacyclovirase: biphenyl hydrolase-like protein as valacyclovir hydrolase
|
|
9801
|
Identification of a human valacyclovirase: biphenyl hydrolase-like protein as valacyclovir hydrolase
|
|
9802
|
Identification of a human valacyclovirase: biphenyl hydrolase-like protein as valacyclovir hydrolase
|
|
9803
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9804
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9805
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9806
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9807
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9808
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9809
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9810
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9811
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9812
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9813
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9814
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9815
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9816
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9817
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9818
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9819
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9820
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9821
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9822
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9823
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9824
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9825
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9826
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9827
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9828
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9829
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9830
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9831
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9832
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9833
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9834
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9835
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9836
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9837
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9838
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9839
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9840
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9841
|
Steady-State Kinetic Characterization of RB69 DNA Polymerase Mutants That Affect dNTP Incorporation
|
|
9842
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9843
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9844
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9845
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9846
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9847
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9848
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9849
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9850
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9851
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9852
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9853
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9854
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9855
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9856
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9857
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9858
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9859
|
Key NAD+-binding residues in human 15-hydroxyprostaglandin dehydrogenase
|
|
9860
|
Cloning, characterization and preliminary crystallographic analysis of Leishmania hypoxanthine-guanine ...
|
|
9861
|
Cloning, characterization and preliminary crystallographic analysis of Leishmania hypoxanthine-guanine ...
|
|
9862
|
Cloning, characterization and preliminary crystallographic analysis of Leishmania hypoxanthine-guanine ...
|
|
9863
|
Cloning, characterization and preliminary crystallographic analysis of Leishmania hypoxanthine-guanine ...
|
|
9864
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9865
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9866
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9867
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9868
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9869
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9870
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9871
|
Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase ...
|
|
9872
|
Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana
|
|
9873
|
Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana
|
|
9874
|
Channelling and formation of `active` formaldehyde in dimethylglycine oxidase
|
|
9875
|
Channelling and formation of `active` formaldehyde in dimethylglycine oxidase
|
|
9876
|
Channelling and formation of `active` formaldehyde in dimethylglycine oxidase
|
|
9877
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9878
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9879
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9880
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9881
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9882
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9883
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9884
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9885
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9886
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9887
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9888
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9889
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9890
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9891
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9892
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9893
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9894
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9895
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9896
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9897
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9898
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9899
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9900
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9901
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9902
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9903
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9904
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9905
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9906
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9907
|
Rapid hepatic metabolism of 7-ketocholesterol by 11beta-hydroxysteroid dehydrogenase type 1: species-specific ...
|
|
9908
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9909
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9910
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9911
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9912
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9913
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9914
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9915
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9916
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9917
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9918
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9919
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9920
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9921
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9922
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9923
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9924
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9925
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9926
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9927
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9928
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9929
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9930
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9931
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9932
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9933
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9934
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9935
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9936
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9937
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9938
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9939
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9940
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9941
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9942
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9943
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9944
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9945
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9946
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9947
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9948
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9949
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9950
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9951
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9952
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9953
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9954
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9955
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9956
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9957
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9958
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9959
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9960
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9961
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9962
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9963
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9964
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9965
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9966
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9967
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9968
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9969
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9970
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9971
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9972
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9973
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9974
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9975
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9976
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9977
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9978
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9979
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9980
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9981
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9982
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9983
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9984
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9985
|
Enzymatic characteristics and subcellular distribution of a short-chain dehydrogenase/reductase family ...
|
|
9986
|
Studies on cystathionine synthase of rat liver. Properties of the highly purified enzyme
|
|
9987
|
Studies on cystathionine synthase of rat liver. Properties of the highly purified enzyme
|
|
9988
|
Studies on cystathionine synthase of rat liver. Properties of the highly purified enzyme
|
|
9989
|
Studies on cystathionine synthase of rat liver. Properties of the highly purified enzyme
|
|
9990
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9991
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9992
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9993
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9994
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9995
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9996
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9997
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9998
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
9999
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|
|
10000
|
Engineering of pyranose 2-oxidase from Peniophora gigantea towards improved thermostability and catalytic ...
|





 If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.
If the current search query is a refinement of the previous query "Use Previous Query (Go Back)" button can be used to perform the previous query again. Only one automatic step back is possible, but the query can be manually manipulated if desired.